5XB0
 
 | |
5XNO
 
 | | Structure of M-LHCII and CP24 complexes in the unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
6KEZ
 
 | | Crystal structure of GAPDH/CP12/PRK complex from Arabidopsis thaliana | | Descriptor: | Calvin cycle protein CP12-2, Glyceraldehyde-3-phosphate dehydrogenase GAPA1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
6KEW
 
 | |
6KEX
 
 | |
6KHJ
 
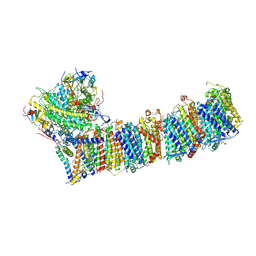 | | Supercomplex for electron transfer | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Pan, X, Cao, D, Xie, F, Zhang, X, Li, M. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Nat Commun, 11, 2020
|
|
6KEV
 
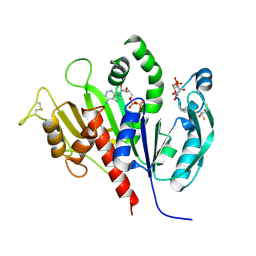 | | Reduced phosphoribulokinase from Synechococcus elongatus PCC 7942 complexed with adenosine diphosphate and glucose 6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yu, A, Xie, Y, Li, M. | | Deposit date: | 2019-07-05 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.506139 Å) | | Cite: | Photosynthetic Phosphoribulokinase Structures: Enzymatic Mechanisms and the Redox Regulation of the Calvin-Benson-Bassham Cycle.
Plant Cell, 32, 2020
|
|
7C9V
 
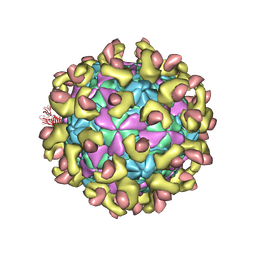 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7DJJ
 
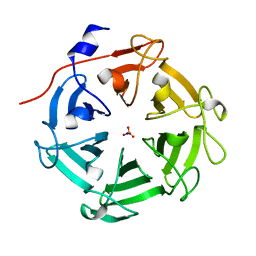 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
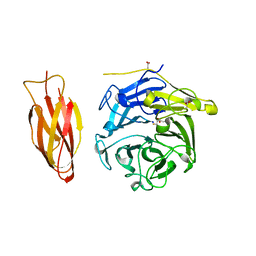 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
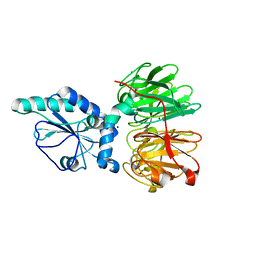 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
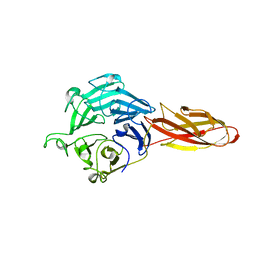 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7CXM
 
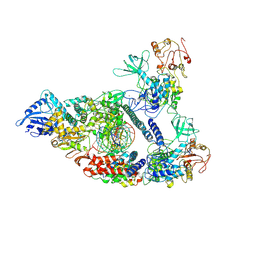 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2K
 
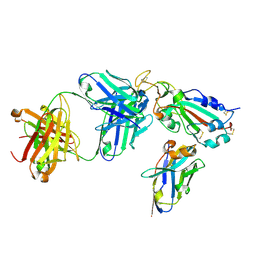 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7RWH
 
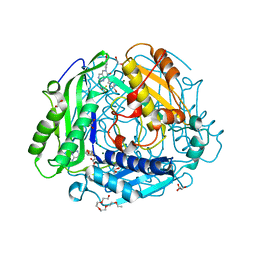 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWG
 
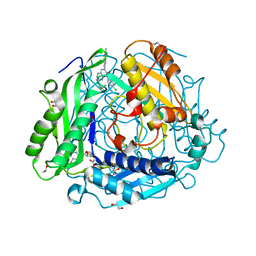 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW7
 
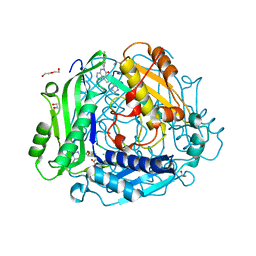 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | Descriptor: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
