6U9L
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM | | Descriptor: | GLYCEROL, POTASSIUM ION, SUBTILISIN BPN', ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6MNL
 
 | |
4RDD
 
 | | Co-crystal structure of SHP2 in complex with a Cefsulodin derivative | | Descriptor: | 1-({(2R)-4-carboxy-2-[(R)-carboxy{[(2R)-2-phenyl-2-sulfoacetyl]amino}methyl]-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyr idinium, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.Y, Yu, Z.H, He, R, Zhang, R.Y. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Exploring the Existing Drug Space for Novel pTyr Mimetic and SHP2 Inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
7KLZ
 
 | |
4N92
 
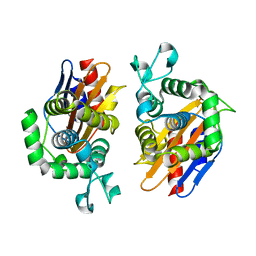 | | Crystal structure of beta-lactamse PenP_E166S | | Descriptor: | Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-19 | | Release date: | 2014-09-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
5ZCX
 
 | | Structure of T20/N39 | | Descriptor: | Envelope glycoprotein, Envelope glycoprotein gp160 | | Authors: | Zhang, X.J, Ding, X.H. | | Deposit date: | 2018-02-21 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional characterization of HIV-1 cell fusion inhibitor T20.
AIDS, 33, 2019
|
|
9JS4
 
 | | Cryo-EM structure of neutralizing antibody 8G3 in complex with BA.1 RBD | | Descriptor: | Heavy chain of 8G3, Light chain of 8G3, Spike glycoprotein | | Authors: | Li, J, Li, H. | | Deposit date: | 2024-09-30 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Rapid restoration of potent neutralization activity against the latest Omicron variant JN.1 via AI rational design and antibody engineering.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4N9L
 
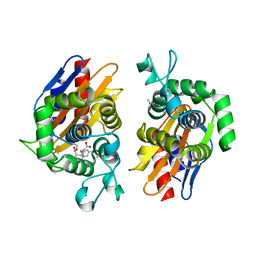 | | crystal structure of beta-lactamse PenP_E166S in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
4N9K
 
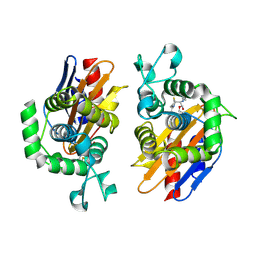 | | crystal structure of beta-lactamse PenP_E166S in complex with cephaloridine | | Descriptor: | 5-METHYL-2-[2-OXO-1-(2-THIOPHEN-2-YL-ACETYLAMINO)-ETHYL]-3,6-DIHYDRO-2H-[1,3]THIAZINE-4-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Pan, X, Wong, W, Zhao, Y. | | Deposit date: | 2013-10-21 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Perturbing the General Base Residue Glu166 in the Active Site of Class A beta-Lactamase Leads to Enhanced Carbapenem Binding and Acylation
Biochemistry, 53, 2014
|
|
6LRD
 
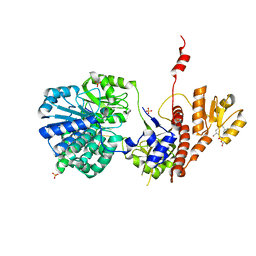 | | Structure of RecJ complexed with a 5'-P-dSpacer-modified ssDNA | | Descriptor: | ASP-LEU-PRO-PHE, DNA (5'-D(P*(3DR)P*TP*TP*TP*TP*T)-3'), MANGANESE (II) ION, ... | | Authors: | Cheng, K, Hua, Y. | | Deposit date: | 2020-01-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901335 Å) | | Cite: | Participation of RecJ in the base excision repair pathway of Deinococcus radiodurans.
Nucleic Acids Res., 48, 2020
|
|
8ZF0
 
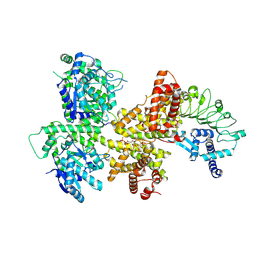 | | pRib-ADP bound OsEDS1-PAD4-ADR1 complex | | Descriptor: | EDS1, Lipase-like PAD4, RPW8 domain-containing protein, ... | | Authors: | Xu, W.Y, Zhang, Y. | | Deposit date: | 2024-05-07 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A canonical protein complex controls immune homeostasis and multipathogen resistance.
Science, 386, 2024
|
|
5GHY
 
 | |
6IUG
 
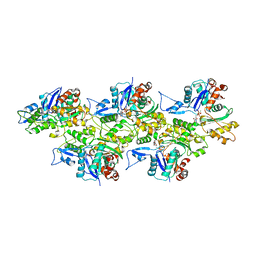 | | Cryo-EM structure of the plant actin filaments from Zea mays pollen | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, pollen F-actin | | Authors: | Ren, Z.H, Zhang, Y, Zhang, Y, He, Y.Q, Du, P.Z, Wang, Z.X, Sun, F, Ren, H.Y. | | Deposit date: | 2018-11-28 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of Actin Filaments fromZea maysPollen.
Plant Cell, 31, 2019
|
|
3NTU
 
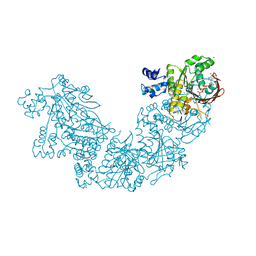 | | RADA RECOMBINASE D302K MUTANT IN COMPLEX with AMP-PNP | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Luo, Y. | | Deposit date: | 2010-07-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RAD51 protein ATP cap regulates nucleoprotein filament stability.
J.Biol.Chem., 287, 2012
|
|
3FV8
 
 | | JNK3 bound to piperazine amide inhibitor, SR2774. | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(3-chloro-2-(4-(prop-2-ynyl)piperazin-1-yl)phenyl)furan-2-carboxamide, Mitogen-activated protein kinase 10 | | Authors: | Habel, J.E. | | Deposit date: | 2009-01-15 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Synthesis and SAR of piperazine amides as novel c-jun N-terminal kinase (JNK) inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8ENX
 
 | |
8ENY
 
 | |
8EO0
 
 | |
8ENZ
 
 | |
8ENS
 
 | |
8ENW
 
 | |
7CO5
 
 | | HtrA-type protease AlgW with decapeptide | | Descriptor: | AlgW protein, IMIDAZOLE, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO3
 
 | | HtrA-type protease AlgWS227A with tripeptide | | Descriptor: | AlgW protein, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO2
 
 | | HtrA-type protease AlgW with tripeptide | | Descriptor: | AlgW protein, IMIDAZOLE, TRP-VAL-PHE | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
7CO7
 
 | | HtrA-type protease AlgWS227A with decapeptide | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, AlgW protein, decapeptide SVRDELRWVF | | Authors: | Li, T, Song, Y.J, Bao, R. | | Deposit date: | 2020-08-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis of the Versatile Regulatory Mechanism of HtrA-Type Protease AlgW from Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
