2NPO
 
 | | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | Acetyltransferase | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative transferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
2NLY
 
 | | Crystal structure of protein BH1492 from Bacillus halodurans, Pfam DUF610 | | Descriptor: | Divergent polysaccharide deacetylase hypothetical protein, ZINC ION | | Authors: | Jin, X, Sauder, J.M, Wasserman, S, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein BH1492 from Bacillus halodurans C-125
To be Published
|
|
2NLZ
 
 | | Crystal structure of cephalosporin acylase from Bacillus halodurans | | Descriptor: | Cephalosporin acylase | | Authors: | Patskovsky, Y, Ramagopal, U, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-20 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cephalosporin acylase from Bacillus halodurans
To be Published
|
|
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
2NRQ
 
 | |
2NV5
 
 | | Crystal structure of a C-terminal phosphatase domain of Rattus norvegicus ortholog of human protein tyrosine phosphatase, receptor type, D (PTPRD) | | Descriptor: | PTPRD, PHOSPHATASE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Iizuka, M, Xu, W, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2NXO
 
 | |
2O56
 
 | | Crystal Structure of a Member of the Enolase Superfamily from Salmonella Typhimurium | | Descriptor: | MAGNESIUM ION, Putative mandelate racemase | | Authors: | Patskovsky, Y, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-05 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Putative Enolase from Salmonella Typhimurium Lt2
To be Published
|
|
2O8R
 
 | | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis | | Descriptor: | Polyphosphate kinase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis
To be Published
|
|
2OGJ
 
 | | Crystal structure of a dihydroorotase | | Descriptor: | Dihydroorotase, IMIDAZOLE, ZINC ION | | Authors: | Sugadev, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of a dihydroorotase
To be Published
|
|
2NRH
 
 | | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni | | Descriptor: | SULFATE ION, Transcriptional activator, putative, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of conserved putative Baf family transcriptional activator from Campylobacter jejuni
To be Published
|
|
2O3J
 
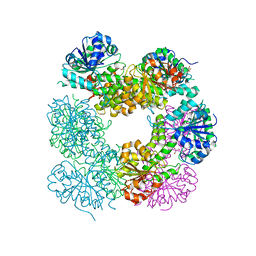 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
2NYG
 
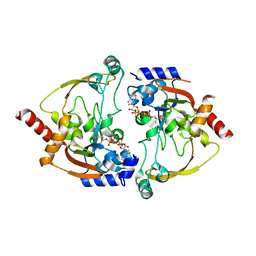 | |
2OHW
 
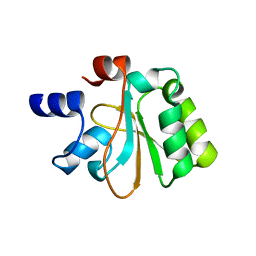 | | Crystal structure of the YueI protein from Bacillus subtilis | | Descriptor: | YueI protein | | Authors: | Bonanno, J.B, Jin, X, Mu, H, Dickey, M, Bain, K.T, Wu, B, Chen, T, Reyes, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-10 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the YueI protein from Bacillus subtilis
To be Published
|
|
2NR4
 
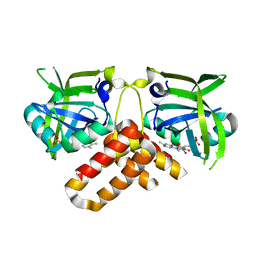 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
2NYV
 
 | | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus | | Descriptor: | Phosphoglycolate phosphatase | | Authors: | Ciatto, C, Min, T, Gorman, J, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | X-ray crystal structure of a phosphoglycolate phosphatase from Aquifex aeolicus
To be Published
|
|
2NWI
 
 | | Crystal structure of protein AF1396 from Archaeoglobus fulgidus, Pfam DUF98 | | Descriptor: | Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Sridhar, V, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-14 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hypothetical protein O28875 from Archaeoglobus fulgidus
To be Published
|
|
2OGF
 
 | | Crystal structure of protein MJ0408 from Methanococcus jannaschii, Pfam DUF372 | | Descriptor: | 8-OXOGUANINE, GLYCEROL, Hypothetical protein MJ0408, ... | | Authors: | Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of hypothetical protein MJ0408 from Methanococcus jannaschii
To be Published
|
|
2NRJ
 
 | | Crystal Structure of Hemolysin binding component from Bacillus cereus | | Descriptor: | Hbl B protein | | Authors: | Madegowda, M, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of the B component of Hemolysin BL from Bacillus cereus
Proteins, 71, 2008
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
2NX2
 
 | | Crystal structure of protein ypsA from Bacillus subtilis, Pfam DUF1273 | | Descriptor: | Hypothetical protein ypsA | | Authors: | Ramagopal, U.A, Alvarado, J, Dickey, M, Reyes, C, Toro, R, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein ypsA from Bacillus subtilis.
To be Published
|
|
2O3A
 
 | | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus | | Descriptor: | UPF0106 protein AF_0751 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Slocombe, A, Sridhar, V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus
To be Published
|
|
2OKT
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
2OG9
 
 | |
