7CDI
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
5Y6P
 
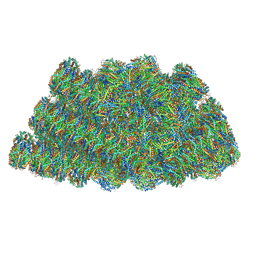 | | Structure of the phycobilisome from the red alga Griffithsia pacifica | | Descriptor: | ApcD, ApcF, LC, ... | | Authors: | Zhang, J, Ma, J.F, Liu, D.S, Sun, S, Sui, S.F. | | Deposit date: | 2017-08-13 | | Release date: | 2017-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of phycobilisome from the red alga Griffithsia pacifica
Nature, 551, 2017
|
|
6KAG
 
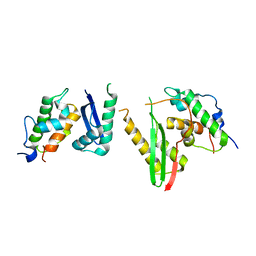 | | Crystal structure of the SMARCB1/SMARCC2 subcomplex | | Descriptor: | SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Chen, G, Zhou, H, Giancotti, F.G, Long, J. | | Deposit date: | 2019-06-22 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | A heterotrimeric SMARCB1-SMARCC2 subcomplex is required for the assembly and tumor suppression function of the BAF chromatin-remodeling complex.
Cell Discov, 6, 2020
|
|
4MK7
 
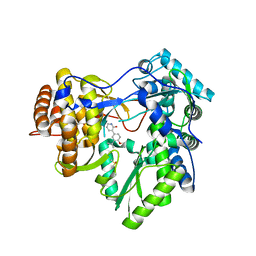 | |
4MK8
 
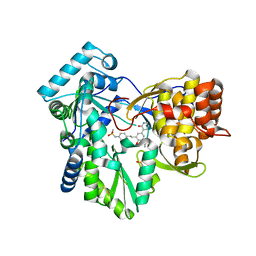 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 4 (N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-(4-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]ethyl}phenyl)methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
4MKA
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 13 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{3-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1-oxo-1H-isochromen-7-yl}methanesulfonamide, ... | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
7E5X
 
 | |
7V42
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 in complex with benzyl-alcohol. | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase, ... | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7V40
 
 | | Crystal structure of Class I P450 monooxygenase (P450tol) from Rhodococcus coprophilus TC-2 | | Descriptor: | PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, p450tol monooxygenase | | Authors: | Zhang, L.L, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2021-08-12 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Molecular Basis for a Toluene Monooxygenase to Govern Substrate Selectivity
Acs Catalysis, 12, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4MKB
 
 | |
4MK9
 
 | | Hepatitis C Virus polymerase NS5B genotype 1b (BK) in complex with inhibitor 12 (N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide) | | Descriptor: | GLYCEROL, N-{2-[3-tert-butyl-2-methoxy-5-(2-oxo-1,2-dihydropyridin-3-yl)phenyl]-1,3-benzoxazol-5-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2013-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a Novel Series of Potent Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B.
J.Med.Chem., 56, 2013
|
|
1BKO
 
 | |
1BSF
 
 | |
1BKP
 
 | |
7W9N
 
 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
5WMT
 
 | |
8WGW
 
 | | Local refinement of RBD-ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wei, X, Zhang, Z. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
