8H03
 
 | |
8H05
 
 | |
8H04
 
 | |
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
6A37
 
 | |
6AW2
 
 | | Crystal structure of the HopQ-CEACAM1 complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AVZ
 
 | | Crystal structure of the HopQ-CEACAM3 WT complex | | Descriptor: | CALCIUM ION, Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AW1
 
 | | Crystal structure of CEACAM3 | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6AW3
 
 | | Crystal structure of the HopQ-CEACAM3 L44Q complex | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 3, HopQ | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
6B0Z
 
 | | IDH1 R132H mutant in complex with IDH305 | | Descriptor: | (4R)-4-[(1S)-1-fluoroethyl]-3-[2-({(1S)-1-[4-methyl-2'-(trifluoromethyl)[3,4'-bipyridin]-6-yl]ethyl}amino)pyrimidin-4-yl]-1,3-oxazolidin-2-one, CITRATE ANION, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2017-09-15 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Discovery and Evaluation of Clinical Candidate IDH305, a Brain Penetrant Mutant IDH1 Inhibitor.
ACS Med Chem Lett, 8, 2017
|
|
6AW0
 
 | | Crystal structure of CEACAM3 L44Q | | Descriptor: | CHLORIDE ION, Carcinoembryonic antigen-related cell adhesion molecule 3, GLYCEROL, ... | | Authors: | Bonsor, D.A, Sundberg, E.J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | TheHelicobacter pyloriadhesin protein HopQ exploits the dimer interface of human CEACAMs to facilitate translocation of the oncoprotein CagA.
EMBO J., 37, 2018
|
|
7W9N
 
 | | THE STRUCTURE OF OBA33-OTA COMPLEX | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, OTA DNA APTAMER (33-MER) | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2021-12-10 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into the Mechanism of High-Affinity Binding of Ochratoxin A by a DNA Aptamer.
J.Am.Chem.Soc., 144, 2022
|
|
7DK3
 
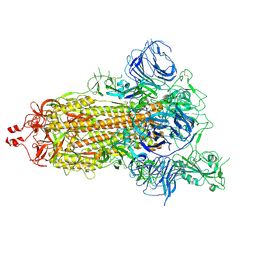 | | SARS-CoV-2 S trimer, S-open | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DD2
 
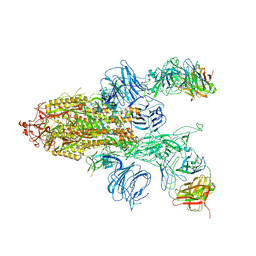 | |
7DK4
 
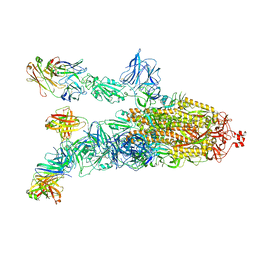 | |
7DDN
 
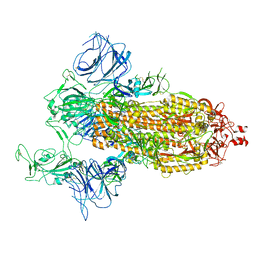 | | SARS-Cov2 S protein at open state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-29 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DF4
 
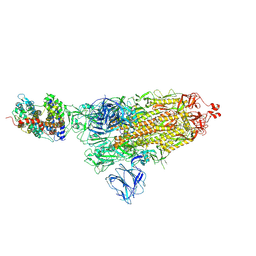 | | SARS-CoV-2 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DDD
 
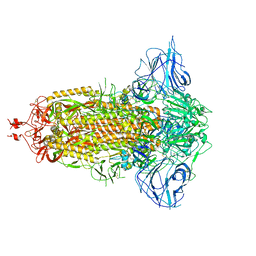 | | SARS-Cov2 S protein at close state | | Descriptor: | Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2020-10-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Development and structural basis of a two-MAb cocktail for treating SARS-CoV-2 infections.
Nat Commun, 12, 2021
|
|
7DF3
 
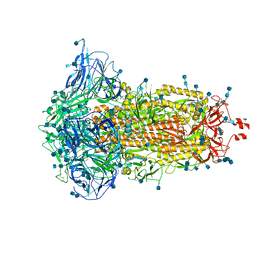 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DCX
 
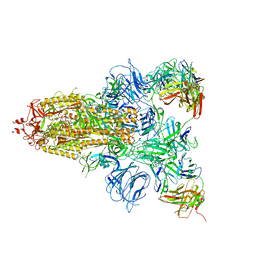 | |
7DK7
 
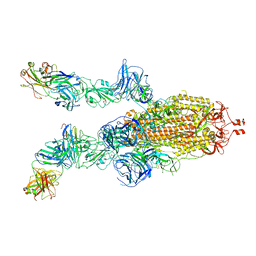 | |
7DCC
 
 | |
