7LIO
 
 | |
7LIQ
 
 | |
6KZ4
 
 | | YebT domain 5-7 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Wang, H.W, Liu, C, Zhang, L. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
7C43
 
 | |
7C4B
 
 | | The crystal structure of Trypanosoma brucei RNase D : UMP complex | | Descriptor: | CCHC-type domain-containing protein, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C4C
 
 | | The crystal structure of Trypanosoma brucei RNase D : GMP complex | | Descriptor: | CCHC-type domain-containing protein, GUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C45
 
 | |
7C47
 
 | | The crystal structure of Trypanosoma brucei RNase D : CMP complex | | Descriptor: | CCHC-type domain-containing protein, CYTIDINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
7C42
 
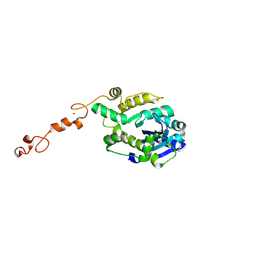 | | The crystal structure of Trypanosoma brucei RNase D | | Descriptor: | CCHC-type domain-containing protein, ZINC ION | | Authors: | Gao, Y.Q, Gan, J.H. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for guide RNA trimming by RNase D ribonuclease in Trypanosoma brucei.
Nucleic Acids Res., 49, 2021
|
|
6KZ3
 
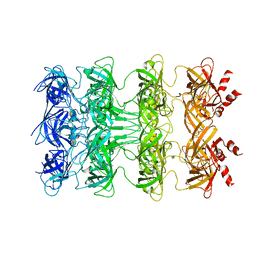 | | YebT domain1-4 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Wang, H.W, Liu, C, Zhang, L. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM Structure of a Bacterial Lipid Transporter YebT.
J.Mol.Biol., 432, 2020
|
|
7FBT
 
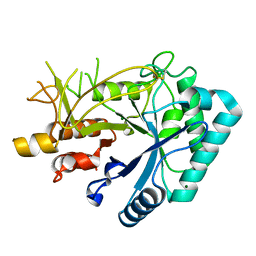 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | Descriptor: | Chitinase, MAGNESIUM ION | | Authors: | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
5FAD
 
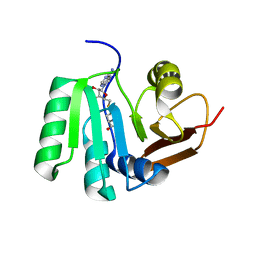 | | SAH complex with aKMT from the hyperthermophilic archaeon Sulfolobus islandicus | | Descriptor: | MAGNESIUM ION, Ribosomal protein L11 methyltransferase, putative, ... | | Authors: | Ouyang, S. | | Deposit date: | 2015-12-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism
Mol.Cell Proteomics, 15, 2016
|
|
5FA8
 
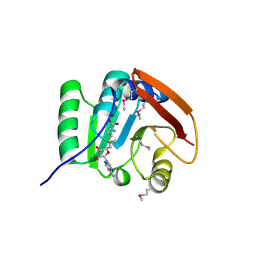 | | SAM complex with aKMT from the hyperthermophilic archaeon Sulfolobus islandicu | | Descriptor: | MAGNESIUM ION, Ribosomal protein L11 methyltransferase, putative, ... | | Authors: | Ouyang, S. | | Deposit date: | 2015-12-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | aKMT Catalyzes Extensive Protein Lysine Methylation in the Hyperthermophilic Archaeon Sulfolobus islandicus but is Dispensable for the Growth of the Organism
Mol.Cell Proteomics, 15, 2016
|
|
6C08
 
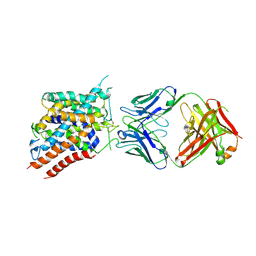 | | Zebrafish SLC38A9 with arginine bound in the cytosol open state | | Descriptor: | ARGININE, Sodium-coupled neutral amino acid transporter 9, antibody Fab Heavy Chain, ... | | Authors: | Lei, H.-T, Gonen, T. | | Deposit date: | 2017-12-28 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2PWL
 
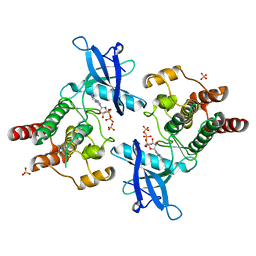 | |
2PVF
 
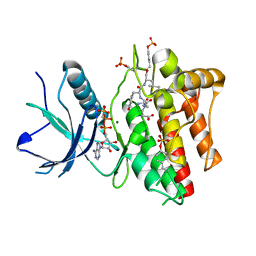 | |
2PZ5
 
 | |
3GZN
 
 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
4JHD
 
 | | Crystal Structure of an Actin Dimer in Complex with the Actin Nucleator Cordon-Bleu | | Descriptor: | Actin-5C, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, X, Ni, F, Wang, Q. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis of actin filament nucleation by tandem w domains.
Cell Rep, 3, 2013
|
|
6INX
 
 | |
7WQG
 
 | | Bovin Alpha-lactalbumin binding with zinc ions | | Descriptor: | Alpha-lactalbumin, ZINC ION | | Authors: | Li, T, Zhang, T. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Zinc binding strength of proteins dominants zinc uptake in Caco-2 cells.
Rsc Adv, 12, 2022
|
|
6S1K
 
 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | Descriptor: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | Authors: | Cassidy, C.K. | | Deposit date: | 2019-06-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.38 Å) | | Cite: | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
7C7X
 
 | |
7YQR
 
 | | Crystal Structure of Xcc NAMPT and its complex with NAM | | Descriptor: | NICOTINAMIDE, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
7YQQ
 
 | | Crystal Structure of Xcc NAMPT and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, PHOSPHATE ION, Pre-B cell enhancing factor related protein | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2022-08-08 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into Xanthomonas campestris pv. campestris NAD + biosynthesis via the NAM salvage pathway.
Commun Biol, 7, 2024
|
|
