9G8K
 
 | | Structure of K+-dependent Na+-PPase from Thermotoga maritima in complex with Ca2+ and Etidronate | | Descriptor: | (1-hydroxyethane-1,1-diyl)bis(phosphonic acid), CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Vidilaseris, K, Liu, J, Goldman, A. | | Deposit date: | 2024-07-23 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Conformational dynamics and asymmetry in multimodal inhibition of membrane-bound pyrophosphatases
Elife, 2024
|
|
9GFY
 
 | | Endothiapepsin in complex with pepstatin soaked at pH 7.6 | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, PENTAETHYLENE GLYCOL, ... | | Authors: | Mueller, J.M, Glinca, S. | | Deposit date: | 2024-08-12 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Protonation Effects in Protein-Ligand Complexes - A Case Study of Endothiapepsin and Pepstatin A with Computational and Experimental Methods.
Chemmedchem, 20, 2025
|
|
9G6U
 
 | | p53-Y220C Core Domain Covalently Bound to 3,5-Dichloro-6-Ethylpyrazine-2-carbonitirle Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 3,5-bis(chloranyl)-6-ethyl-pyrazine-2-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | SNAr Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes
Drug Des Devel Ther, 2025
|
|
9G6T
 
 | | p53-Y220C Core Domain Covalently Bound to 5-Chloro-6-methylpyrazine-2-carbonitrile Soaked at 5 mM | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-chloranyl-6-methyl-pyrazine-2-carbonitrile, ... | | Authors: | Stahlecker, J, Klett, T, Stehle, T, Boeckler, F.M. | | Deposit date: | 2024-07-19 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | SNAr Reactive Pyrazine Derivatives as p53-Y220C Cleft Binders with Diverse Binding Modes
Drug Des Devel Ther, 2025
|
|
7AYG
 
 | | oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7B2E
 
 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7ABB
 
 | |
7A1A
 
 | |
9H6E
 
 | |
9H3I
 
 | | trans-aconitate decarboxylase Tad1- wild type | | Descriptor: | Trans-aconitate decarboxylase 1 | | Authors: | Zheng, L, Bang, G. | | Deposit date: | 2024-10-16 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanistic and structural insights into the itaconate-producing trans -aconitate decarboxylase Tad1.
Pnas Nexus, 4, 2025
|
|
9H4G
 
 | |
9H4H
 
 | | trans-aconitate decarboxylase Tad1_S320A | | Descriptor: | Trans-aconitate decarboxylase 1 | | Authors: | Zheng, L, Bange, G. | | Deposit date: | 2024-10-18 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Mechanistic and structural insights into the itaconate-producing trans -aconitate decarboxylase Tad1.
Pnas Nexus, 4, 2025
|
|
9H4E
 
 | |
9EOI
 
 | |
9EU4
 
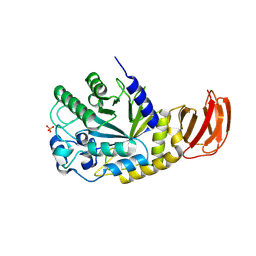 | | GH29A alpha-L-fucosidase | | Descriptor: | Exported alpha-L-fucosidase protein, SULFATE ION, beta-L-fucopyranose | | Authors: | Yang, Y.Y, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-03-27 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural elucidation and characterization of GH29A alpha-l-fucosidases and the effect of pH on their transglycosylation.
Febs J., 292, 2025
|
|
2FR2
 
 | |
7OUI
 
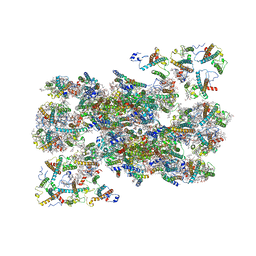 | | Structure of C2S2M2-type Photosystem supercomplex from Arabidopsis thaliana (digitonin-extracted) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Graca, A.T, Hall, M, Persson, K, Schroder, W.P. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | High-resolution model of Arabidopsis Photosystem II reveals the structural consequences of digitonin-extraction.
Sci Rep, 11, 2021
|
|
7OR2
 
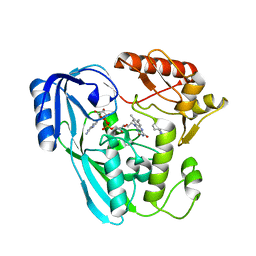 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 4) | | Descriptor: | 5-methyl-1-phenyl-pyrazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-04 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OSQ
 
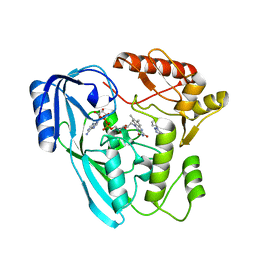 | | Crystal structure of UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) from Pseudomonas aeruginosa in complex with FAD and a pyrazole derivative (fragment 18) | | Descriptor: | 5-methyl-1-phenyl-1,2,3-triazole-4-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, UDP-N-acetylenolpyruvoylglucosamine reductase | | Authors: | Acebron-Garcia de Eulate, M, Mayol-Llinas, J, Blundell, T.L, Kim, S.Y, Mendes, V, Abell, C. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Novel Inhibitors of Uridine Diphosphate- N -Acetylenolpyruvylglucosamine Reductase (MurB) from Pseudomonas aeruginosa , an Opportunistic Infectious Agent Causing Death in Cystic Fibrosis Patients.
J.Med.Chem., 65, 2022
|
|
7OS2
 
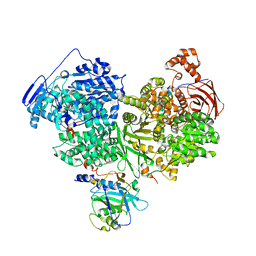 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS1
 
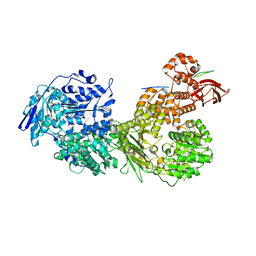 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OOR
 
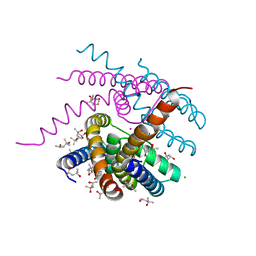 | | NaK C-DI mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OPH
 
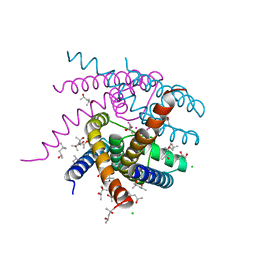 | | NaK S-DI mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-31 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OOU
 
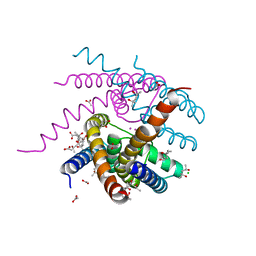 | | NaK C-DI mutant with Li+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7OQ2
 
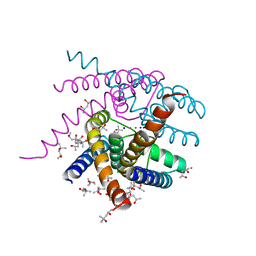 | | NaK S-DI mutant soaked in Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
