5JYY
 
 | | Structure-based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-resistant Influenza Viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-[(2-methoxyethyl)carbamoyl]-D-glycero-D-galacto-non-2-enon ic acid, CALCIUM ION, ... | | Authors: | Fu, L, Wu, Y, Bi, Y, Zhang, S, Lv, X, Qi, J, Li, Y, Lu, X, Yan, J, Gao, G.F, Li, X. | | Deposit date: | 2016-05-15 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Tetravalent Zanamivir with Potent Inhibitory Activity against Drug-Resistant Influenza Viruses
J.Med.Chem., 59, 2016
|
|
4WVD
 
 | | Identification of a novel FXR ligand that regulates metabolism | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, Bile acid receptor, FORMIC ACID, ... | | Authors: | Wang, R, Li, Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The antiparasitic drug ivermectin is a novel FXR ligand that regulates metabolism.
Nat Commun, 4, 2013
|
|
6WSL
 
 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
1L4U
 
 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AND PT(II) AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | Deposit date: | 2002-03-05 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
6VYC
 
 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
1L4Y
 
 | | CRYSTAL STRUCTURE OF SHIKIMATE KINASE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MGADP AT 2.0 ANGSTROM RESOLUTION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Gu, Y, Reshetnikova, L, Li, Y, Wu, Y, Yan, H, Singh, S, Ji, X. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of shikimate kinase from Mycobacterium tuberculosis reveals the dynamic role of the LID domain in catalysis.
J.Mol.Biol., 319, 2002
|
|
8HD5
 
 | |
4H53
 
 | | Influenza N2-Tyr406Asp neuraminidase in complex with beta-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
6KNW
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
6KNV
 
 | | THRb mutation with a novel agonist | | Descriptor: | 2-[(1-methyl-4-oxidanyl-7-phenoxy-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-08-07 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Revealing a Mutant-Induced Receptor Allosteric Mechanism for the Thyroid Hormone Resistance.
Iscience, 20, 2019
|
|
8AS8
 
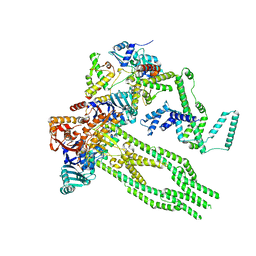 | | E. coli Wadjet JetABC monomer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Li, Y, Myasnikov, A, Gruber, S. | | Deposit date: | 2022-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
8A9W
 
 | | Crystal structure of PulL C-ter domain | | Descriptor: | SULFATE ION, Type II secretion system protein L | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8AB1
 
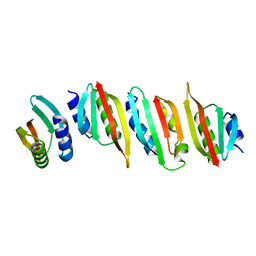 | | Crystal structure of the PulL-PulM C-terminal domain heterocomplex | | Descriptor: | Type II secretion system protein L, Type II secretion system protein M | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8A9X
 
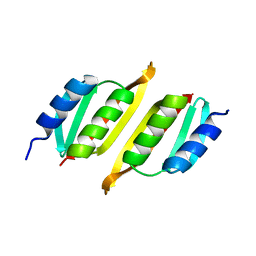 | | Crystal structure of PulM C-ter domain | | Descriptor: | Type II secretion system protein M | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
3MD1
 
 | | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1) | | Descriptor: | GLYCEROL, Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Li, H, Shi, H, Li, Y, Cui, Y, Niu, L, Teng, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Second RRM Domain of Yeast Poly(U)-Binding Protein (Pub1)
To be published
|
|
5MSZ
 
 | | Lytic Polysaccharide Monooxygenase AA15 from Thermobia domestica in the Cu(I) State | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Thermobia domestica domestica AA15 | | Authors: | Hemsworth, G.R, Sabbadin, F, Ciano, L, Henrissat, B, Dupree, P, Tryfona, T, Besser, K, Elias, L, Pesante, G, Li, Y, Dowle, A, Bates, R, Gomez, L, Hallam, R, Davies, G.J, Walton, P.H, Bruce, N.C, McQueen-Mason, S. | | Deposit date: | 2017-01-06 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An ancient family of lytic polysaccharide monooxygenases with roles in arthropod development and biomass digestion.
Nat Commun, 9, 2018
|
|
8BFN
 
 | | E. coli Wadjet JetABC dimer of dimers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Myasnikov, A, Li, Y, Gruber, S. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
4H52
 
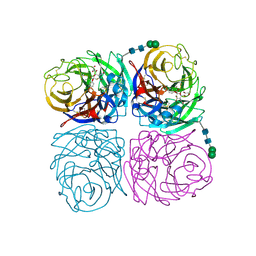 | | Wild-type influenza N2 neuraminidase covalent complex with 3-fluoro-Neu5Ac | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-3,5-dideoxy-3-fluoro-D-erythro-alpha-L-manno-non-2-ulopyranosonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Liu, Y, Kiyota, H, Sriwilaijaroen, N, Qi, J, Tanaka, K, Wu, Y, Li, Q, Li, Y, Yan, J, Suzuki, Y, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Influenza neuraminidase operates via a nucleophilic mechanism and can be targeted by covalent inhibitors
Nat Commun, 4, 2013
|
|
8BS6
 
 | |
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
3R45
 
 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
1Z2E
 
 | | Solution Structure of Bacillus subtilis ArsC in oxidized state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
8T9A
 
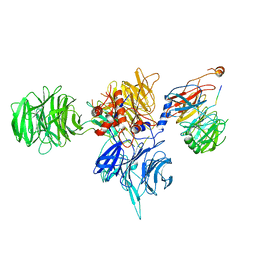 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | Authors: | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Probing the CRL4 DCAF12 interactions with MAGEA3 and CCT5 di-Glu C-terminal degrons.
Pnas Nexus, 3, 2024
|
|
1Z2D
 
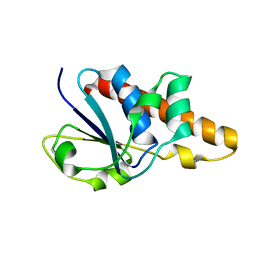 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
