1XFR
 
 | |
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1X7B
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X76
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor 1 (alpha), [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1XBX
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, 5-O-phosphono-L-ribulose, MAGNESIUM ION, ... | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1ZGW
 
 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1YEY
 
 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
2A43
 
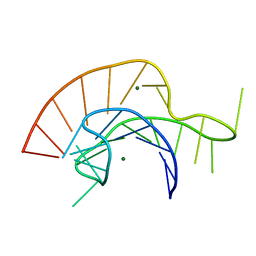 | | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif | | Descriptor: | MAGNESIUM ION, RNA Pseudoknot | | Authors: | Pallan, P.S, Marshall, W.S, Harp, J, Jewett III, F.C, Wawrzak, Z, Brown II, B.A, Rich, A, Egli, M. | | Deposit date: | 2005-06-27 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of a Luteoviral RNA Pseudoknot and Model for a Minimal Ribosomal Frameshifting Motif
Biochemistry, 44, 2005
|
|
7KQN
 
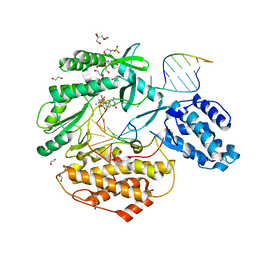 | | Ternary complex of TERT (telomerase reverse transcriptase) with RNA template, DNA primer, an incoming dGTP and a downstream hybrid duplex | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7KQM
 
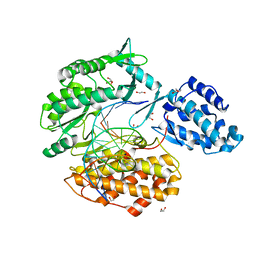 | | Binary complex of TERT (telomerase reverse transcriptase) with RNA/telomeric DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*GP*CP*AP*CP*CP*TP*G)-3'), ... | | Authors: | Choi, W.S, Weng, P.J, Yang, W. | | Deposit date: | 2020-11-17 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7LBX
 
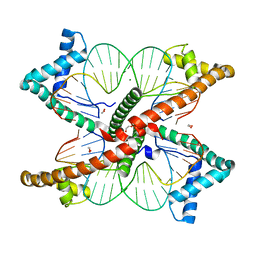 | |
7LBW
 
 | |
7KYQ
 
 | |
7KYS
 
 | | Crystal structure of human BCCIP beta (Native2) | | Descriptor: | 1,2-ETHANEDIOL, BRCA2 and CDKN1A-interacting protein, DI(HYDROXYETHYL)ETHER | | Authors: | Choi, W.S, Liu, B, Shen, Z, Yang, W. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human BCCIP and implications for binding and modification of partner proteins.
Protein Sci., 30, 2021
|
|
5XLJ
 
 | | Crystal structure of the flagellar cap protein flid D2-D3 domains from serratia marcescens in Space group P432 | | Descriptor: | CHLORIDE ION, Flagellar hook-associated protein 2, SODIUM ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
5Y6H
 
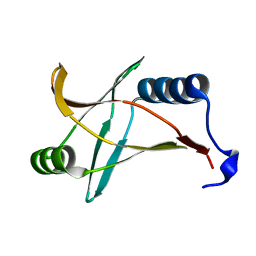 | |
5YHH
 
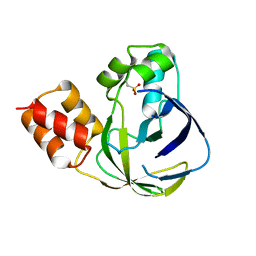 | | Crystal structure of YiiM from Geobacillus stearothermophilus | | Descriptor: | Uncharacterized conserved protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5YHI
 
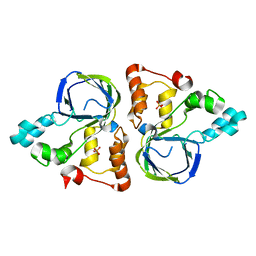 | | Crystal structure of YiiM from Escherichia coli | | Descriptor: | PHOSPHATE ION, Protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5Y8T
 
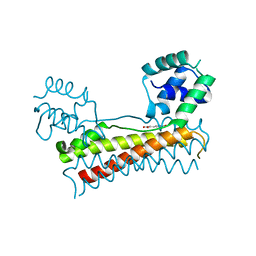 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
5X11
 
 | | Crystal structure of Bacillus subtilis PadR in complex with operator DNA | | Descriptor: | DNA (28-MER), Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X14
 
 | | Crystal structure of Bacillus subtilis PadR in complex with ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5ZIY
 
 | | Crystal structure of Bacillus cereus FlgL | | Descriptor: | Flagellar hook-associated protein 3, ZINC ION | | Authors: | Hong, H.J, Kim, T.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2018-03-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FlgL and its implications for flagellar assembly
Sci Rep, 8, 2018
|
|
5Z2L
 
 | | Crystal structure of BdcA in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | Authors: | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | Deposit date: | 2018-01-03 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5ZIZ
 
 | |
