7RYW
 
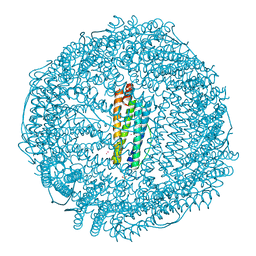 | | Crystal structure of Ferritin grown by the microbatch method in the presence of Agarose | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, D.J, Stojanoff, V. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Ferritin grown by microbatch method in the presence of agarose
To Be Published
|
|
7RZX
 
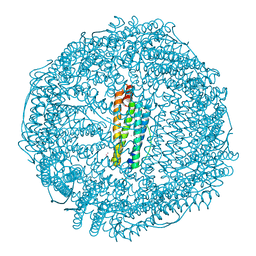 | | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, J.D, Stojanoff, V. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV
To Be Published
|
|
4OO8
 
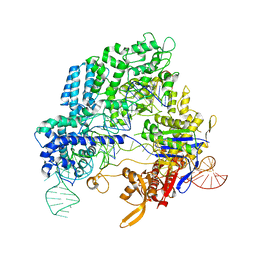 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
3WTR
 
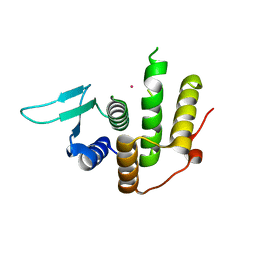 | | Crystal structure of E. coli YfcM bound to Co(II) | | Descriptor: | COBALT (II) ION, Uncharacterized protein | | Authors: | Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-19 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation
Nucleic Acids Res., 42, 2014
|
|
3WUH
 
 | | Qri7 and AMP complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, tRNA N6-adenosine threonylcarbamoyltransferase, ... | | Authors: | Tominaga, T, Kobayashi, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2014-04-24 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.937 Å) | | Cite: | Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP
ACTA CRYSTALLOGR.,SECT.F, 70, 2014
|
|
8GOS
 
 | | Crystal structure of fluorescent protein RasM | | Descriptor: | RasM | | Authors: | Adachi, M, Kagotani, Y, Shimizu, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Beat-frequency-resolved two-dimensional electronic spectroscopy: disentangling vibrational coherences in artificial fluorescent proteins with sub-10-fs visible laser pulses.
Opt Express, 31, 2023
|
|
5ZN4
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148N mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN0
 
 | | Joint X-ray/neutron structure of protein kinase ck2 alpha subunit | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Ostermann, A, Schrader, T.E, Sunami, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN1
 
 | | X-ray structure of protein kinase ck2 alpha subunit in D2O | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN5
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148A mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN3
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148S mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5ZN2
 
 | | X-ray structure of protein kinase ck2 alpha subunit H148A mutant | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION | | Authors: | Shibazaki, C, Arai, S, Shimizu, R, Kinoshita, T, Kuroki, R, Adachi, M. | | Deposit date: | 2018-04-07 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydration Structures of the Human Protein Kinase CK2 alpha Clarified by Joint Neutron and X-ray Crystallography.
J. Mol. Biol., 430, 2018
|
|
5AWW
 
 | | Precise Resting State of Thermus thermophilus SecYEG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
6A27
 
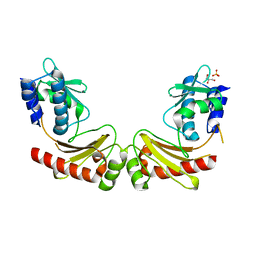 | | Crystal structure of PprA W183R mutant form 1 | | Descriptor: | DNA repair protein PprA, GLYCEROL, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
6A29
 
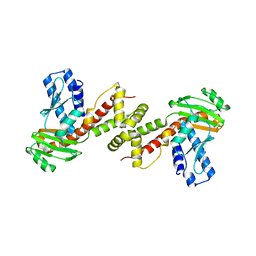 | | Crystal structure of PprA A139R mutant | | Descriptor: | DNA repair protein PprA | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
5CH4
 
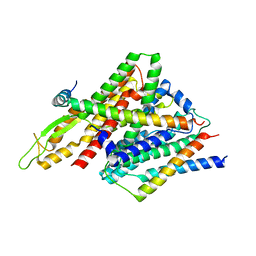 | | Peptide-Bound State of Thermus thermophilus SecYEG | | Descriptor: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | Authors: | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | Deposit date: | 2015-07-10 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5GVS
 
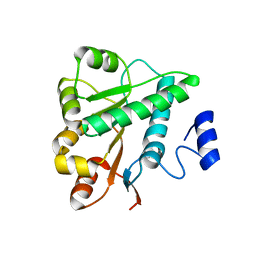 | | Crystal structure of the DDX41 DEAD domain in an apo open form | | Descriptor: | Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
5GVR
 
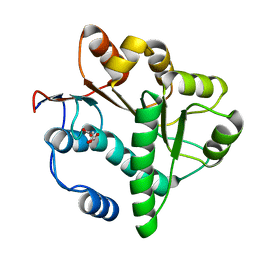 | | Crystal structure of the DDX41 DEAD domain in an apo closed form | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Probable ATP-dependent RNA helicase DDX41 | | Authors: | Omura, H, Oikawa, D, Nakane, T, Kato, M, Ishii, R, Goto, Y, Suga, H, Ishitani, R, Tokunaga, F, Nureki, O. | | Deposit date: | 2016-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Analysis of DDX41: a bispecific immune receptor for DNA and cyclic dinucleotide
Sci Rep, 6, 2016
|
|
6A28
 
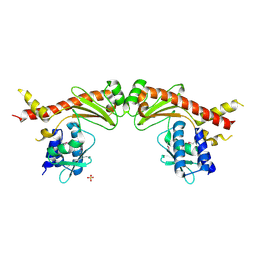 | | Crystal structure of PprA W183R mutant form 2 | | Descriptor: | DNA repair protein PprA, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
7DPA
 
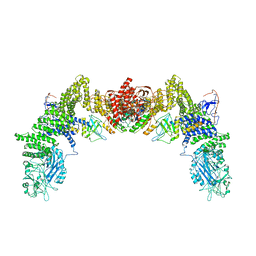 | | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Kaushik, R, Ehara, H, Yokoyama, T, Uchikubo-Kamo, T, Mishima-Tsumagari, C, Yonemochi, M, Ikeda, M, Hanada, K, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Sci Adv, 7, 2021
|
|
8HUK
 
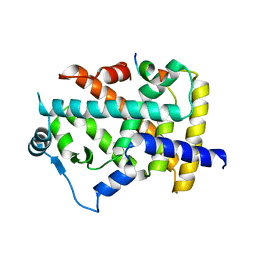 | | X-ray structure of human PPAR alpha ligand binding domain-lanifibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 4-[1-(1,3-benzothiazol-6-ylsulfonyl)-5-chloro-indol-2-yl]butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
8HUQ
 
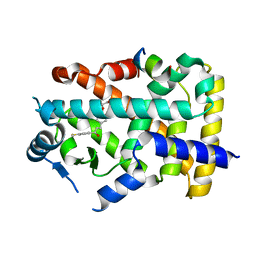 | | X-ray structure of human PPAR alpha ligand binding domain-elafibranor-SRC1 coactivator peptide co-crystals obtained by soaking | | Descriptor: | 15-meric peptide from Nuclear receptor coactivator 1, 2-[2,6-dimethyl-4-[(~{E})-3-(4-methylsulfanylphenyl)-3-oxidanylidene-prop-1-enyl]phenoxy]-2-methyl-propanoic acid, GLYCEROL, ... | | Authors: | Kamata, S, Ishikawa, R, Akahane, M, Honda, A, Oyama, T, Ishii, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional and Structural Insights into the Human PPAR alpha / delta / gamma Targeting Preferences of Anti-NASH Investigational Drugs, Lanifibranor, Seladelpar, and Elafibranor.
Antioxidants, 12, 2023
|
|
5ZSU
 
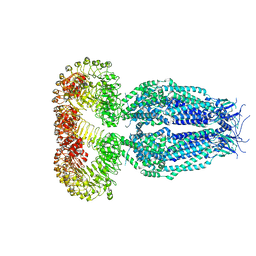 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3WS4
 
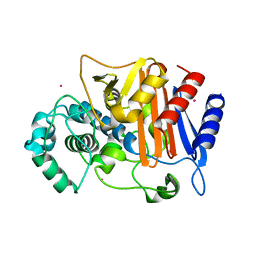 | | N288Q-N321Q mutant BETA-LACTAMASE DERIVED FROM CHROMOHALOBACTER SP.560 (Condition-2A) | | Descriptor: | Beta-lactamase, CHLORIDE ION, STRONTIUM ION | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WRT
 
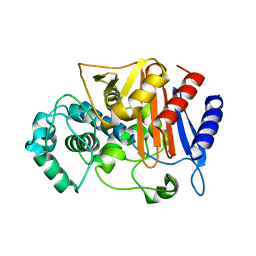 | | Wild type beta-lactamase DERIVED FROM CHROMOHALOBACTER SP.560 | | Descriptor: | Beta-lactamase | | Authors: | Arai, S, Yonezawa, Y, Okazaki, N, Matsumoto, F, Shimizu, R, Yamada, M, Adachi, M, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a highly acidic beta-lactamase from the moderate halophile Chromohalobacter sp. 560 and the discovery of a Cs(+)-selective binding site
Acta Crystallogr.,Sect.D, 71, 2015
|
|
