1A4P
 
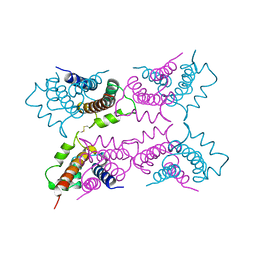 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
1BT6
 
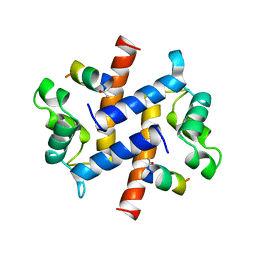 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
3ZYP
 
 | | Cellulose induced protein, Cip1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CIP1, ... | | Authors: | Jacobson, F, Karkehabadi, S, Hansson, H, Goedegebuur, F, Wallace, L, Mitchinson, C, Piens, K, Stals, I, Sandgren, M. | | Deposit date: | 2011-08-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of the Core Domain of a Cellulose Induced Protein (Cip1) from Hypocrea Jecorina, at 1.5 A Resolution.
Plos One, 8, 2013
|
|
1ML5
 
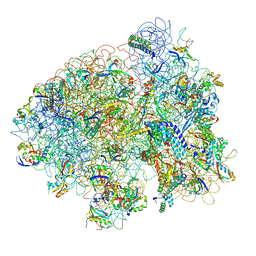 | | Structure of the E. coli ribosomal termination complex with release factor 2 | | Descriptor: | 30S 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Klaholz, B.P, Pape, T, Zavialov, A.V, Myasnikov, A.G, Orlova, E.V, Vestergaard, B, Ehrenberg, M, van Heel, M. | | Deposit date: | 2002-08-30 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structure of the Escherichia coli ribosomal termination complex with release factor 2
Nature, 421, 2003
|
|
1MI6
 
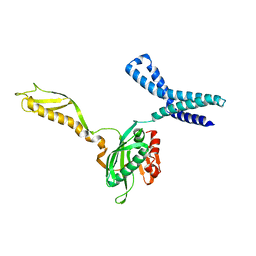 | | Docking of the modified RF2 X-ray structure into the Low Resolution Cryo-EM map of RF2 E.coli 70S Ribosome | | Descriptor: | peptide chain release factor RF-2 | | Authors: | Rawat, U.B.S, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
4BXT
 
 | |
4CS8
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 2 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CS7
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer, form 1 | | Descriptor: | M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
4CS9
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to adenosine monophosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, M2-1, ZINC ION | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
2CDQ
 
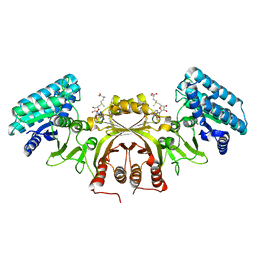 | | Crystal structure of Arabidopsis thaliana aspartate kinase complexed with lysine and S- adenosylmethionine | | Descriptor: | ASPARTOKINASE, D(-)-TARTARIC ACID, LYSINE, ... | | Authors: | Mas-Droux, C, Curien, G, Robert-Genthon, M, Laurencin, M, Ferrer, J.L, Dumas, R. | | Deposit date: | 2006-01-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Organization of Act Domains in Allosteric Enzymes Revealed by the Crystal Structure of Arabidopsis Aspartate Kinase
Plant Cell, 18, 2006
|
|
2CKR
 
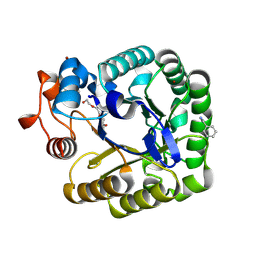 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) E355Q IN COMPLEX WITH CELLOTETRAOSE | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
2CL2
 
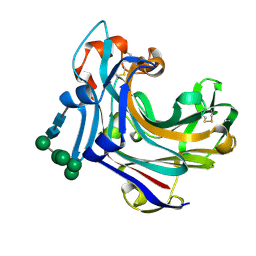 | | Endo-1,3(4)-beta-glucanase from Phanerochaete chrysosporium, solved using native sulfur SAD, exhibiting intact heptasaccharide glycosylation | | Descriptor: | PUTATIVE LAMINARINASE, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vasur, J, Kawai, R, Igarashi, K, Sandgren, M, Samejima, M, Stahlberg, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray crystallographic native sulfur SAD structure determination of laminarinase Lam16A from Phanerochaete chrysosporium.
Acta Crystallogr. D Biol. Crystallogr., 62, 2006
|
|
1QZD
 
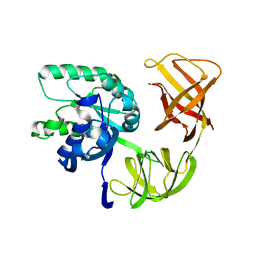 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1R2X
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1PN6
 
 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | Descriptor: | Elongation factor G | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
2CKS
 
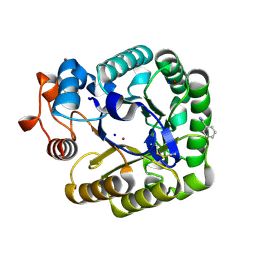 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THERMOBIFIDA FUSCA ENDOGLUCANASE CEL5A (E5) | | Descriptor: | BENZAMIDINE, ENDOGLUCANASE E-5, SODIUM ION, ... | | Authors: | Berglund, G.I, Gualfetti, P.J, Requadt, C, Gross, L.S, Bergfors, T, Shaw, A, Saldajeno, M, Mitchinson, C, Sandgren, M. | | Deposit date: | 2006-04-21 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Catalytic Domain of Thermobifida Fusca Endoglucanase Cel5A in Complex with Cellotetraose
To be Published
|
|
1PN7
 
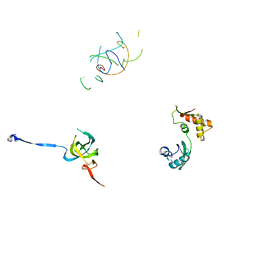 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1R2W
 
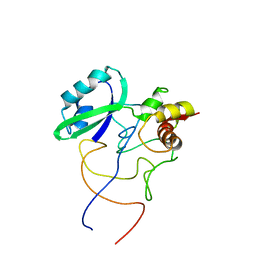 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
2FOM
 
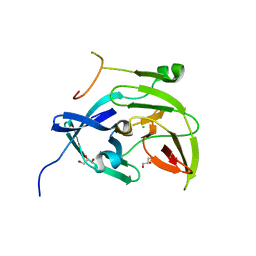 | | Dengue Virus NS2B/NS3 Protease | | Descriptor: | CHLORIDE ION, GLYCEROL, polyprotein | | Authors: | Schiering, N, Kroemer, M, Renatus, M, Erbel, P, D'Arcy, A. | | Deposit date: | 2006-01-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1PN8
 
 | | Coordinates of S12, L11 proteins and E-site tRNA from 70S crystal structure separately fitted into the Cryo-EM map of E.coli 70S.EF-G.GDPNP complex. The atomic coordinates originally from the E-site tRNA were fitted in the position of the hybrid P/E-site tRNA. | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, E-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
7KCR
 
 | | Cryo-EM structure of Zika virus in complex with E protein cross-linking human monoclonal antibody ADI30056 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADI30056 Fab heavy chain variable region, ADI30056 Fab light chain variable region, ... | | Authors: | Sevvana, M, Rogers, T.F, Miller, A.S, Long, F, Klose, T, Beutler, N, Lai, Y.C, Parren, M, Walker, L.M, Buda, G, Burton, D.R, Rossmann, M.G, Kuhn, R.J. | | Deposit date: | 2020-10-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections.
Viruses, 12, 2020
|
|
1QZC
 
 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1YRS
 
 | | Crystal structure of KSP in complex with inhibitor 1 | | Descriptor: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
