3VS2
 
 | | Crystal structure of HCK complexed with a pyrrolo-pyrimidine inhibitor 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine | | Descriptor: | 7-[cis-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Tomabechi, Y, Niwa, H, Parker, J.L, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
3NBS
 
 | | Crystal structure of dimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4E9T
 
 | | Multicopper Oxidase CueO (data6) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-03-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site.
J.Mol.Biol., 373, 2007
|
|
3EOQ
 
 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | Descriptor: | Putative zinc protease | | Authors: | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-09-29 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
1ON0
 
 | | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SR144 | | Descriptor: | CHLORIDE ION, SULFATE ION, YycN protein | | Authors: | Forouhar, F, Shen, J, Kuzin, A, Chiang, Y, Xiao, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-02-26 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Putative Acetyltransferase (YycN) from Bacillus subtilis
To be Published
|
|
2E8I
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, D1 mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Shibata, N, Higuchi, Y, Negoro, S. | | Deposit date: | 2007-01-20 | | Release date: | 2008-01-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold.
Febs J., 276, 2009
|
|
5XVB
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an H2-reduced condition | | Descriptor: | FE3-S4 CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
1UBJ
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
6LS8
 
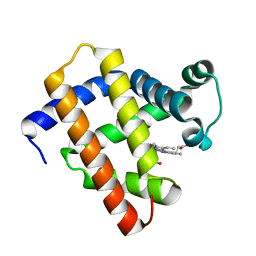 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
5Y34
 
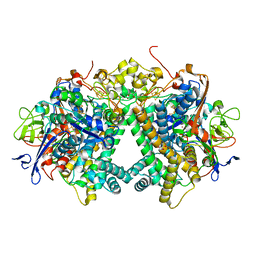 | | Membrane-bound respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus in a ferricyanide-oxidized condition | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
6LTM
 
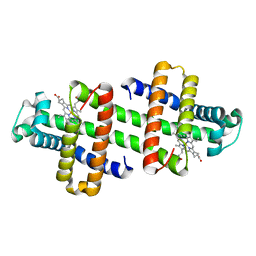 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTL
 
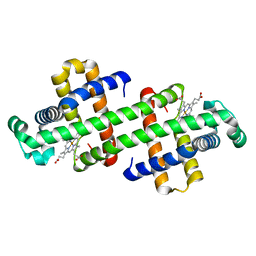 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
8JVC
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVG
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
8JVF
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
3QHE
 
 | | Crystal structure of the complex between the armadillo repeat domain of adenomatous polyposis coli and the tyrosine-rich domain of Sam68 | | Descriptor: | Adenomatous polyposis coli protein, KH domain-containing, RNA-binding, ... | | Authors: | Morishita, E.C.J, Murayama, K, Kato-Murayama, M, Ishizuku-Katsura, Y, Tomabechi, Y, Terada, T, Handa, N, Shirouzu, M, Akiyama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-01-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the armadillo repeat domain of adenomatous polyposis coli and its complex with the tyrosine-rich domain of sam68
Structure, 19, 2011
|
|
3RFN
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
4LM5
 
 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | Descriptor: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5Z25
 
 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
3RHU
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | SC_1wnu | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3RI0
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_2cx5_001, GLYCEROL, SULFATE ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3RIJ
 
 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | GLYCEROL, SC_2cx5 | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
