3WH3
 
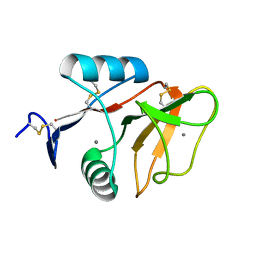 | | human Mincle, ligand free form | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WAR
 
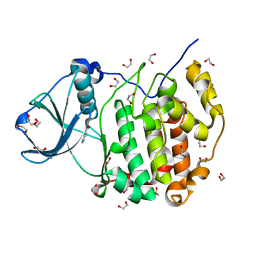 | | Crystal structure of human CK2a | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, NICOTINIC ACID | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Sogabe, Y, Sakurai, A, Nakamura, S, Nakanishi, I. | | Deposit date: | 2013-05-07 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structure of human CK2 alpha at 1.06 angstrom resolution
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3WOW
 
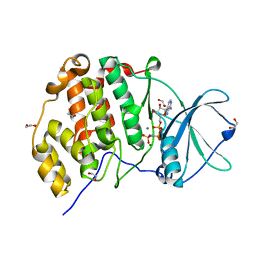 | | Crystal structure of human CK2a with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | Authors: | Kinoshita, T, Nakaniwa, T, Sekiguchi, Y, Sogabe, Y, Sakurai, A, Nakamura, S, Nakanishi, I, Shimada, K, Tanaka, M. | | Deposit date: | 2014-01-06 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic residue divergence of CK2a contribute to a species-dependent variation for apigenin binding mode but not for an ATP analogue
To be Published
|
|
3U2J
 
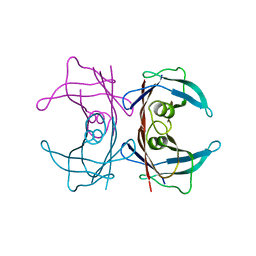 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3U2I
 
 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3VV0
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with DAAM-3 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][2-(hexylamino)ethyl]amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2E8J
 
 | |
2DLC
 
 | | Crystal structure of the ternary complex of yeast tyrosyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, O-(ADENOSINE-5'-O-YL)-N-(L-TYROSYL)PHOSPHORAMIDATE, T-RNA (76-MER), ... | | Authors: | Tsunoda, M, Kusakabe, Y, Tanaka, N, Nakamura, K.T. | | Deposit date: | 2006-04-18 | | Release date: | 2007-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms.
Nucleic Acids Res., 35, 2007
|
|
5XQZ
 
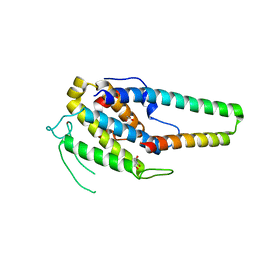 | | Structure of the MOB1-NDR2 complex | | Descriptor: | GLYCEROL, MOB kinase activator 1A, Serine/threonine-protein kinase 38-like, ... | | Authors: | Wu, G, Lin, K. | | Deposit date: | 2017-06-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stable MOB1 interaction with Hippo/MST is not essential for development and tissue growth control.
Nat Commun, 8, 2017
|
|
5B13
 
 | | Crystal structure of phycoerythrin | | Descriptor: | PHYCOCYANOBILIN, PHYCOUROBILIN, Phycoerythrin alpha subunit, ... | | Authors: | Tanaka, Y, Gai, Z, Kishimura, H. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural properties of phycoerythrin from dulse palmaria palmata
J FOOD BIOCHEM., 2016
|
|
3ATG
 
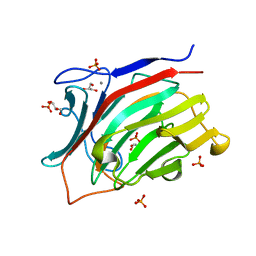 | | endo-1,3-beta-glucanase from Cellulosimicrobium cellulans | | Descriptor: | CALCIUM ION, GLUCANASE, GLYCEROL, ... | | Authors: | Tanabe, Y, Pang, Z, Oda, M, Mikami, B. | | Deposit date: | 2011-01-04 | | Release date: | 2012-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and thermodynamic characterization of endo-1,3-beta-glucanase: Insights into the substrate recognition mechanism.
Biochim. Biophys. Acta, 1866, 2018
|
|
1KXW
 
 | |
2RPP
 
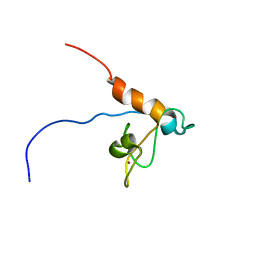 | | Solution structure of Tandem zinc finger domain 12 in Muscleblind-like protein 2 | | Descriptor: | Muscleblind-like protein 2, ZINC ION | | Authors: | Abe, C, Dang, W, Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-06-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
3WO8
 
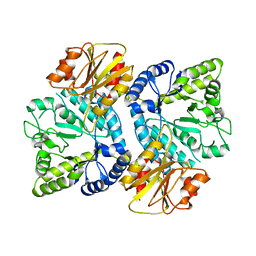 | | Crystal structure of the beta-N-acetylglucosaminidase from Thermotoga maritima | | Descriptor: | Beta-N-acetylglucosaminidase | | Authors: | Mine, S, Kado, Y, Watanabe, M, Inoue, T, Ishikawa, K. | | Deposit date: | 2013-12-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The structure of hyperthermophilic beta-N-acetylglucosaminidase reveals a novel dimer architecture associated with the active site.
Febs J., 281, 2014
|
|
8IG0
 
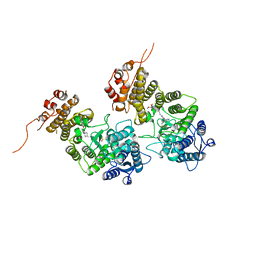 | | Crystal structure of menin in complex with DS-1594b | | Descriptor: | (1R,2S,4R)-4-[[4-(5,6-dimethoxypyridazin-3-yl)phenyl]methylamino]-2-[methyl-[6-[2,2,2-tris(fluoranyl)ethyl]thieno[2,3-d]pyrimidin-4-yl]amino]cyclopentan-1-ol, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Suzuki, M, Yoneyama, T, Imai, E. | | Deposit date: | 2023-02-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel Menin-MLL1 inhibitor, DS-1594a, prevents the progression of acute leukemia with rearranged MLL1 or mutated NPM1.
Cancer Cell Int, 23, 2023
|
|
1KXX
 
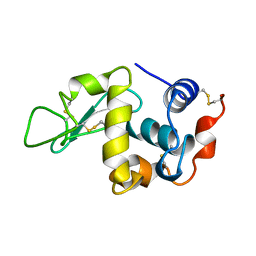 | |
1KXY
 
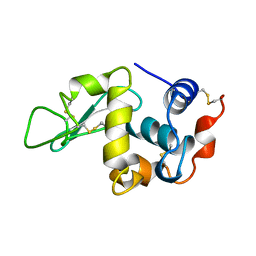 | |
1RFP
 
 | |
3WL4
 
 | | N,N'-diacetylchitobiose deacetylase (Se-derivative) from Pyrococcus furiosus | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Uegaki, K. | | Deposit date: | 2013-11-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Expression from engineered Escherichia coli chromosome and crystallographic study of archaeal N,N'-diacetylchitobiose deacetylase
Febs J., 281, 2014
|
|
5YY4
 
 | | Crystal structure of the scFv antibody 4B08 with sulfated epitope peptide | | Descriptor: | C-C chemokine receptor type 5, CHLORIDE ION, SULFATE ION, ... | | Authors: | Caaveiro, J.M.M, Miyanabe, K, Tsumoto, K. | | Deposit date: | 2017-12-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Tyrosine Sulfation Restricts the Conformational Ensemble of a Flexible Peptide, Strengthening the Binding Affinity for an Antibody
Biochemistry, 57, 2018
|
|
2E5S
 
 | | Solution structure of the zf-CCCHx2 domain of muscleblind-like 2, isoform 1 [Homo sapiens] | | Descriptor: | OTTHUMP00000018578, ZINC ION | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA binding domain in the human muscleblind-like protein 2
Protein Sci., 18, 2009
|
|
7VGP
 
 | |
7VGO
 
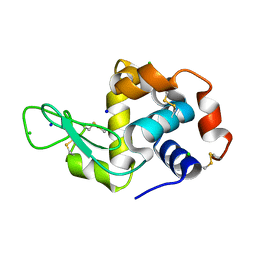 | | Hen egg lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Oda, M, Ikura, T, Ito, N. | | Deposit date: | 2021-09-17 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Hen Egg Lysozyme Refolded after Denaturation at Acidic pH.
Protein J., 41, 2022
|
|
2ZTB
 
 | |
