8OHB
 
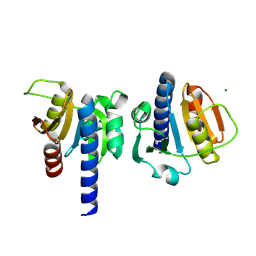 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry E08 | | Descriptor: | 5-(2-methoxyethyl)-1,3,4-oxadiazol-2-amine, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OHC
 
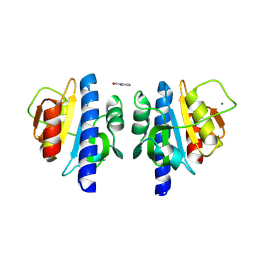 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry E12 | | Descriptor: | 6-azanyl-3-methyl-1,3-benzoxazol-2-one, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OHG
 
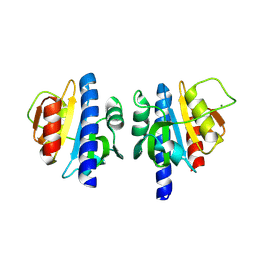 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F09 | | Descriptor: | 4-pyridin-2-ylphenol, Cyclic di-AMP synthase CdaA, MAGNESIUM ION | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8OHE
 
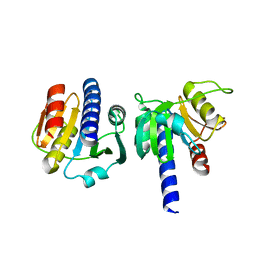 | | PanDDA analysis group deposition -- CdaA in complex with fragment F2X-Entry F03 | | Descriptor: | Cyclic di-AMP synthase CdaA, MAGNESIUM ION, N-(3-fluorophenyl)-2-(2-methoxyethoxy)acetamide | | Authors: | Garbers, T.B, Neumann, P, Wollenhaupt, J, Weiss, M.S, Ficner, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-04-03 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic fragment screen of the c-di-AMP-synthesizing enzyme CdaA from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2O0R
 
 | | The three-dimensional structure of N-Succinyldiaminopimelate aminotransferase from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, GLYCEROL, Rv0858c (N-Succinyldiaminopimelate aminotransferase), ... | | Authors: | Weyand, S, Kefala, G, Weiss, M.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-11-28 | | Release date: | 2007-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Three-dimensional Structure of N-Succinyldiaminopimelate Aminotransferase from Mycobacterium tuberculosis
J.Mol.Biol., 367, 2007
|
|
2O0T
 
 | |
2Y41
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with IPM and MN | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y3Z
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - apo enzyme | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-ISOPROPYLMALATE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y40
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, MANGANESE (II) ION | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
2Y42
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus - complex with NADH and Mn | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, BICINE, MANGANESE (II) ION, ... | | Authors: | Graczer, E, merlin, A, Singh, R.K, Manikandan, K, Zavodsky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2011-01-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Level Description of the Domain Closure in a Dimeric Enzyme: Thermus Thermophilus 3-Isopropylmalate Dehydrogenase.
Mol.Biosyst., 7, 2011
|
|
1YAZ
 
 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
9RCZ
 
 | |
4F7I
 
 | | Structure of Isopropylmalate dehydrogenase from Thermus thermophilus in complex with IPM, Mn and NADH | | Descriptor: | 3-ISOPROPYLMALIC ACID, 3-isopropylmalate dehydrogenase, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Pallo, A, Graczer, E, Zavodszky, P, Weiss, M.S, Vas, M. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and energetic basis of isopropylmalate dehydrogenase enzyme catalysis.
Febs J., 281, 2014
|
|
5J5Z
 
 | | Crystal structure of the D444V disease-causing mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Mizsei, R, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2016-04-04 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of the disease-causing D444V mutant and the relevant wild type human dihydrolipoamide dehydrogenase.
Free Radic. Biol. Med., 124, 2018
|
|
4ZE6
 
 | | Endothiapepsin in complex with fragment B39 | | Descriptor: | 1,2-ETHANEDIOL, 7-aminoheptanoic acid, ACETATE ION, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-04-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3HFW
 
 | | Crystal Structure of human ADP-ribosylhydrolase 1 (hARH1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mueller-Dieckmann, C, Weiss, M.S, Mueller-Dieckmann, J, Koch-Nolte, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of human ADP-ribosylhydrolase 1
To be Published
|
|
4ZEA
 
 | | Endothiapepsin in complex with fragment B91 | | Descriptor: | 1,2-ETHANEDIOL, 2-IMINOBIOTIN, ACETATE ION, ... | | Authors: | Huschmann, F.U, Linnik, J, Weiss, M.S, Mueller, U. | | Deposit date: | 2015-04-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of endothiapepsin-fragment complexes from crystallographic fragment screening using a novel, diverse and affordable 96-compound fragment library.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4OL4
 
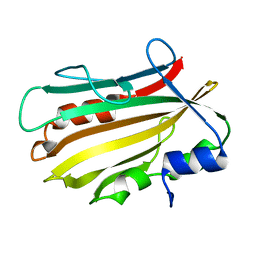 | | Crystal structure of secreted proline rich antigen MTC28 (Rv0040c) from Mycobacterium tuberculosis | | Descriptor: | Proline-rich 28 kDa antigen | | Authors: | Kundu, P, Biswas, R, Mukherjee, S, Reinhard, L, Mueller-dieckmann, J, Weiss, M.S, Das, A.K. | | Deposit date: | 2014-01-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Epitope Mapping of Mycobacterium tuberculosis Secretary Antigen MTC28
J.Biol.Chem., 291, 2016
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2VK8
 
 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant E477Q in complex with its substrate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-17 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
2Q27
 
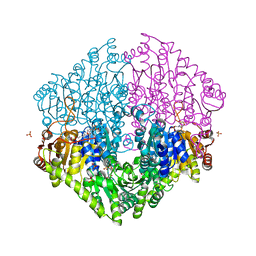 | | Crystal structure of oxalyl-coA decarboxylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Werther, T, Zimmer, A, Wille, G, Hubner, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli.
Febs J., 277, 2010
|
|
2XYO
 
 | | Structural basis for a new tetracycline resistance mechanism relying on the TetX monooxygenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Volkers, G, Palm, G.J, Weiss, M.S, Hinrichs, W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2R8P
 
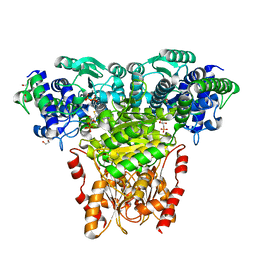 | | Transketolase from E. coli in complex with substrate D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium-2-yl}-6-O-phosphono-D-glucitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
2R8O
 
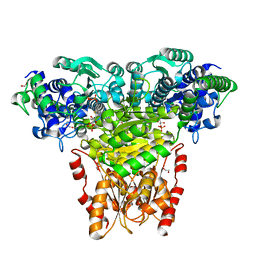 | | Transketolase from E. coli in complex with substrate D-xylulose-5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thia zol-3-ium-2-yl}-5-O-phosphono-D-xylitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
1JCV
 
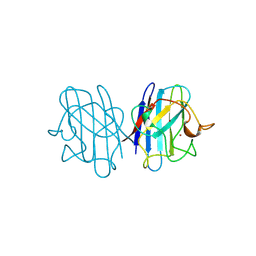 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE LOW TEMPERATURE (-180C) STRUCTURE | | Descriptor: | COPPER (II) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Ogihara, N.L, Parge, H.E, Hart, P.J, Weiss, M.S, Valentine, J.S, Eisenberg, D.S, Tainer, J.A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
