3AS8
 
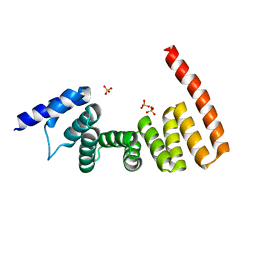 | | MamA MSR-1 P41212 | | Descriptor: | Magnetosome protein MamA, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ASH
 
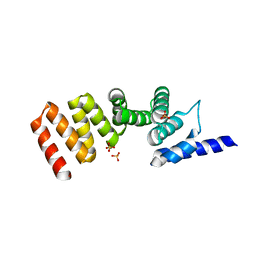 | | MamA D159K mutant 1 | | Descriptor: | MamA, SULFATE ION | | Authors: | Zeytuni, N, Levin, M, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ASG
 
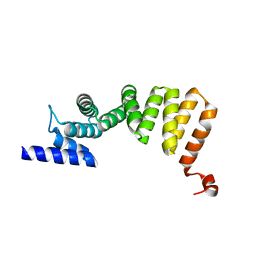 | | MamA D159K mutant 2 | | Descriptor: | MamA | | Authors: | Zeytuni, N, Levin, M, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1HPB
 
 | |
2RDV
 
 | |
5COK
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-0476 | | Descriptor: | (3aS,4S,7aR)-hexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COP
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-097 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-(4-methoxyphenyl)butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5CON
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-015 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
5COO
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
1MJ5
 
 | | LINB (haloalkane dehalogenase) from sphingomonas paucimobilis UT26 at atomic resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Oakley, A.J, Damborsky, J, Wilce, M.C. | | Deposit date: | 2002-08-27 | | Release date: | 2003-08-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26 at 0.95 A resolution: dynamics of catalytic residues.
Biochemistry, 43, 2004
|
|
1RDV
 
 | |
3FR0
 
 | | Human glucokinase in complex with 2-amino benzamide activator | | Descriptor: | 2-amino-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of novel and potent 2-amino benzamide derivatives as allosteric glucokinase activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1IZ7
 
 | |
1IZ8
 
 | | Re-refinement of the structure of hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | Descriptor: | 1,3-PROPANDIOL, BROMIDE ION, CALCIUM ION, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
1MPG
 
 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
5C9I
 
 | |
1IRY
 
 | | Solution structure of the hMTH1, a nucleotide pool sanitization enzyme | | Descriptor: | hMTH1 | | Authors: | Mishima, M, Itoh, N, Sakai, Y, Kamiya, H, Nakabeppu, Y, Shirakawa, M. | | Deposit date: | 2001-10-25 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of human MTH1, a Nudix family hydrolase that selectively degrades oxidized purine nucleoside triphosphates
J.Biol.Chem., 279, 2004
|
|
4YFA
 
 | |
4YFB
 
 | |
4YF9
 
 | |
5MXA
 
 | | Structure of unbound Interleukin-23 | | Descriptor: | Interleukin-12 subunit beta, Interleukin-23 subunit alpha, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-01-22 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
6LFW
 
 | |
3GOI
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 2-(methylamino)-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, alpha-D-glucopyranose | | Authors: | Kamata, K, Mitsuya, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of novel 3,6-disubstituted 2-pyridinecarboxamide derivatives as GK activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6LFV
 
 | |
5Y6T
 
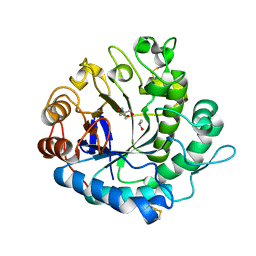 | | Crystal structure of endo-1,4-beta-mannanase from Eisenia fetida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, endo-1,4-beta-mannanase | | Authors: | Hirano, Y, Ueda, M, Tamada, T. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gene cloning, expression, and X-ray crystallographic analysis of a beta-mannanase from Eisenia fetida.
Enzyme.Microb.Technol., 117, 2018
|
|
