3UQP
 
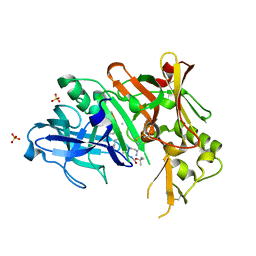 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7W8S
 
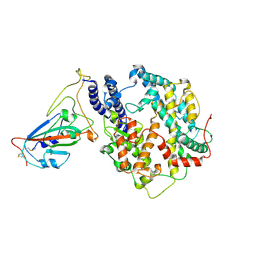 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
 | |
6LNZ
 
 | |
6M31
 
 | |
7F26
 
 | | Crystal structure of lysozyme | | Descriptor: | Lysozyme C | | Authors: | Liang, M. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single pulse data collection with an X-ray chopper at in situ room temperature Laue crystallography beamline BL03HB
Nucl Instrum Methods Phys Res A, 1069, 2024
|
|
7V3T
 
 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
2LKG
 
 | | WSA major conformation | | Descriptor: | Acetylcholine receptor | | Authors: | Xu, Y, Mowrey, D, Cui, T, Perez-Aguilar, J.M, Saven, J.G, Eckenhoff, R, Tang, P. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor.
Biochim.Biophys.Acta, 1818, 2011
|
|
7BPU
 
 | |
8I8A
 
 | |
8I8C
 
 | |
8I8B
 
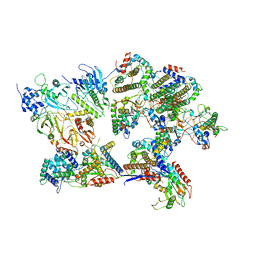 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
8IAZ
 
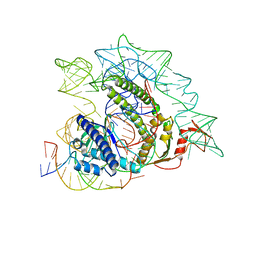 | | Cryo-EM structure of the ISFba1 TnpB-reRNA-dsDNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*AP*TP*GP*GP*AP*CP*CP*AP*TP*CP*AP*GP*CP*TP*CP*CP*TP*AP*AP*TP*GP*G)-3'), DNA (5'-D(P*CP*CP*AP*TP*TP*AP*GP*GP*AP*GP*CP*TP*GP*AP*TP*G)-3'), RNA (207-MER), ... | | Authors: | Yin, M, Zhou, F, Zhu, Y, Huang, Z. | | Deposit date: | 2023-02-09 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Discovery and structural mechanism of DNA endonucleases guided by RAGATH-18-derived RNAs.
Cell Res., 34, 2024
|
|
5YEJ
 
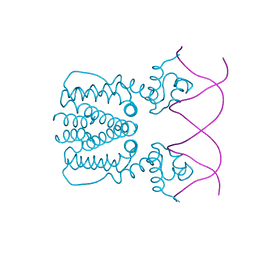 | | Crystal structure of BioQ with its naturel double-stranded DNA operator | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*AP*AP*CP*AP*CP*CP*GP*TP*TP*CP*AP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*TP*GP*AP*AP*CP*GP*GP*TP*GP*TP*TP*CP*AP*GP*GP*T)-3'), TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
5YEK
 
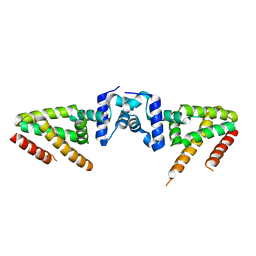 | | Crystal structure of BioQ | | Descriptor: | TetR family transcriptional regulator | | Authors: | Yan, L, Guan, Z.Y, Zou, T.T. | | Deposit date: | 2017-09-17 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural insights into operator recognition by BioQ in the Mycobacterium smegmatis biotin synthesis pathway.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7C6Q
 
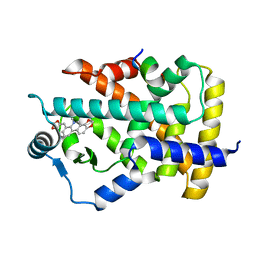 | | Novel natural PPARalpha agonist with a unique binding mode | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Peroxisome proliferator-activated receptor alpha | | Authors: | Tian, S.Y, Wang, R, Zheng, W.L, Li, Y. | | Deposit date: | 2020-05-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Basis for PPARs Activation by The Dual PPAR alpha / gamma Agonist Sanguinarine: A Unique Mode of Ligand Recognition.
Molecules, 26, 2021
|
|
7CLF
 
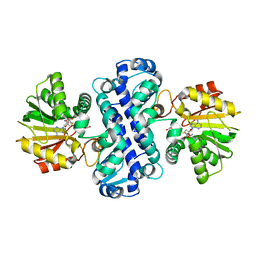 | | PigF with SAH | | Descriptor: | ACETATE ION, Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
5Y5R
 
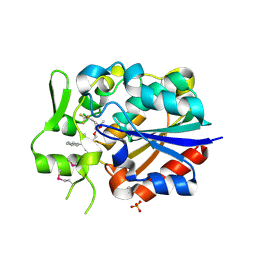 | | Crystal structure of a novel Pyrethroid Hydrolase PytH with BIF | | Descriptor: | (2-methyl-3-phenyl-phenyl)methyl (1~{S})-3-[(~{E})-2-chloranyl-3,3,3-tris(fluoranyl)prop-1-enyl]-2,2-dimethyl-cyclopropane-1-carboxylate, Pyrethroid hydrolase, SULFATE ION | | Authors: | Xu, D.Q, Ran, T.T, Wang, W.W. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-15 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure and Catalytic Mechanism of a Novel Pyrethroid Hydrolase from Sphingobium faniae JZ-2
To Be Published
|
|
7BWQ
 
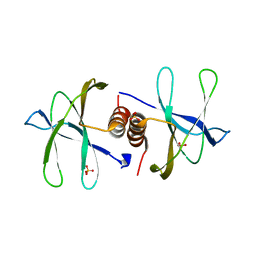 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
7CLU
 
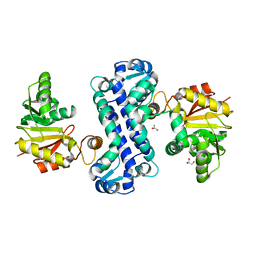 | | PigF with SAH | | Descriptor: | ACETATE ION, GLYCEROL, Methyltransferase domain-containing protein | | Authors: | Qiu, S, Xu, D, Han, N, Sun, B, Ran, T, Wang, W. | | Deposit date: | 2020-07-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of PigF, an O-methyltransferase involved in the prodigiosin synthetic pathway, reveal an induced-fit substrate-recognition mechanism.
Iucrj, 9, 2022
|
|
7EFT
 
 | |
7DCD
 
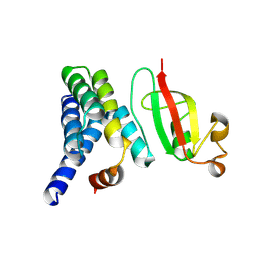 | |
5ZJK
 
 | | Structure of myroilysin | | Descriptor: | Myroilysin, PHOSPHATE ION, ZINC ION | | Authors: | Li, W.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of mature myroilysin and implication for its activation mechanism.
Int.J.Biol.Macromol., 2019
|
|
7V4Y
 
 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
