6EMA
 
 | |
1WO5
 
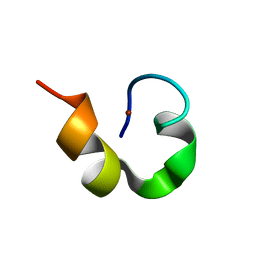 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
6ERT
 
 | |
6ERV
 
 | |
1AQT
 
 | |
2YMD
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with serotonin (5-hydroxytryptamine) | | Descriptor: | GLYCEROL, PHOSPHATE ION, SEROTONIN, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht(3) Receptors.
Embo Rep., 14, 2013
|
|
4EDV
 
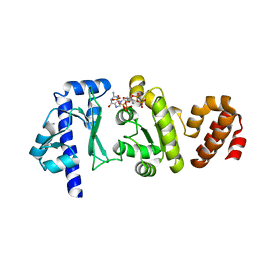 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to pppGpp and Manganese | | Descriptor: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | Authors: | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | Deposit date: | 2012-03-27 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
1X3W
 
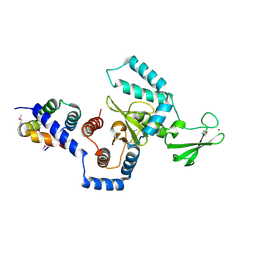 | | Structure of a peptide:N-glycanase-Rad23 complex | | Descriptor: | UV excision repair protein RAD23, ZINC ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, ... | | Authors: | Lee, J.-H, Choi, J.M, Lee, C, Yi, K.J, Cho, Y. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a peptide:N-glycanase-Rad23 complex: insight into the deglycosylation for denatured glycoproteins.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WLR
 
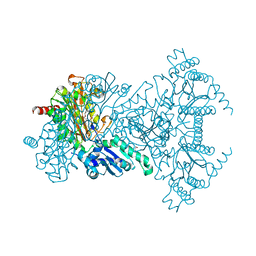 | | Apo aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, TETRAETHYLENE GLYCOL, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-06-29 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional implications of metal ion selection in aminopeptidase p, a metalloprotease with a dinuclear metal center
Biochemistry, 44, 2005
|
|
2Z3S
 
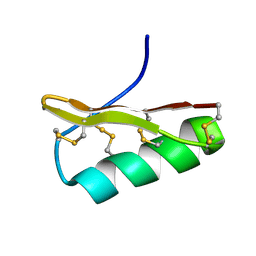 | | NMR structure of AgTx2-MTX | | Descriptor: | AgTx2-MTX | | Authors: | Pimentel, C, M'Barrek, S, Visan, V, Grissmer, S, Sabatier, J.M, Darbon, H, Fajloun, Z. | | Deposit date: | 2007-06-06 | | Release date: | 2008-04-22 | | Last modified: | 2020-02-26 | | Method: | SOLUTION NMR | | Cite: | Chemical synthesis and 1H-NMR 3D structure determination of AgTx2-MTX chimera, a new potential blocker for Kv1.2 channel, derived from MTX and AgTx2 scorpion toxins.
Protein Sci., 17, 2008
|
|
2Z8O
 
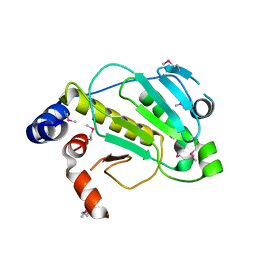 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, L(+)-TARTARIC ACID | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4EPM
 
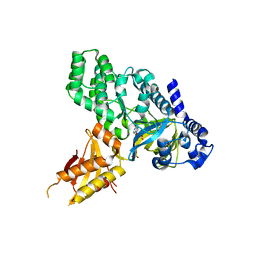 | | Crystal Structure of Arabidopsis GH3.12 (PBS3) in Complex with AMP | | Descriptor: | 4-substituted benzoates-glutamate ligase GH3.12, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Westfall, C.S, Zubieta, C, Herrmann, J, Kapp, U, Nanao, M.H, Jez, J.M. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for prereceptor modulation of plant hormones by GH3 proteins.
Science, 336, 2012
|
|
2ZL7
 
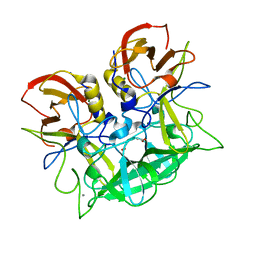 | | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus | | Descriptor: | 58 kd capsid protein, ACETATE ION, CALCIUM ION, ... | | Authors: | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2INX
 
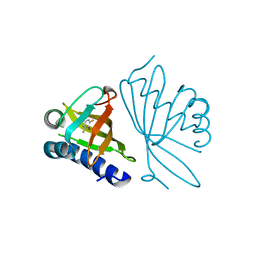 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
4EER
 
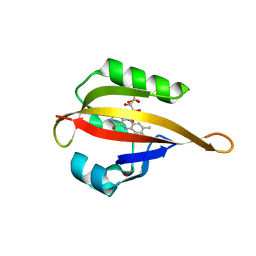 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
2ZIM
 
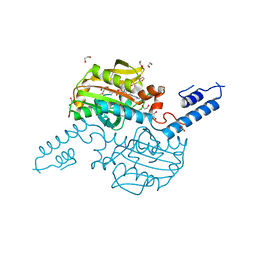 | | Pyrrolysyl-tRNA synthetase bound to adenylated pyrrolysine and pyrophosphate | | Descriptor: | (2R)-2-AMINO-6-({[(2S,3R)-3-METHYLPYRROLIDIN-2-YL]CARBONYL}AMINO)HEXANOYL [(2S,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL HYDROGEN (R)-PHOSPHATE, 1,2-ETHANEDIOL, PYROPHOSPHATE 2-, ... | | Authors: | Steitz, T.A, Kavran, J.M. | | Deposit date: | 2008-02-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZL5
 
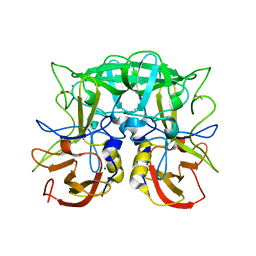 | | Atomic resolution structural characterization of recognition of histo-blood group antigen by Norwalk virus | | Descriptor: | 58 kd capsid protein, ACETATE ION, CALCIUM ION, ... | | Authors: | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1WL9
 
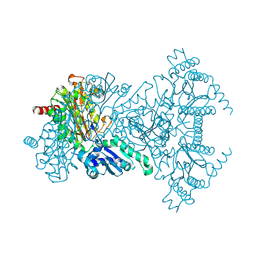 | | Structure of aminopeptidase P from E. coli | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Xaa-Pro aminopeptidase | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-06-22 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional implications of metal ion selection in aminopeptidase p, a metalloprotease with a dinuclear metal center
Biochemistry, 44, 2005
|
|
2ZL6
 
 | | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus | | Descriptor: | 58 kd capsid protein, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Choi, J.M, Huston, A.M, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2008-04-02 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Atomic resolution structural characterization of recognition of histo-blood group antigens by Norwalk virus
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4CO6
 
 | | Crystal structure of the Nipah virus RNA free nucleoprotein- phosphoprotein complex | | Descriptor: | BROMIDE ION, CHLORIDE ION, NUCLEOPROTEIN, ... | | Authors: | Yabukarksi, F, Lawrence, P, Tarbouriech, N, Bourhis, J.M, Jensen, M.R, Ruigrok, R.W.H, Blackledge, M, Volchkov, V, Jamin, M. | | Deposit date: | 2014-01-27 | | Release date: | 2014-08-13 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure of Nipah Virus Unassembled Nucleoprotein in Complex with its Viral Chaperone.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4D9S
 
 | | Crystal structure of Arabidopsis thaliana UVR8 (UV Resistance locus 8) | | Descriptor: | UVB-resistance protein UVR8 | | Authors: | Arvai, A.S, Christie, J.M, Pratt, A.J, Hitomi, K, Getzoff, E.D. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Science, 335, 2012
|
|
4D0T
 
 | | GalNAc-T2 crystal soaked with UDP-GalNAc, EA2 peptide and manganese | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, MANGANESE (II) ION, ... | | Authors: | Lira-Navarrete, E, Iglesias-Fernandez, J, Zandberg, W.F, Companon, I, Kong, Y, Corzana, F, Pinto, B.M, Clausen, H, Peregrina, J.M, Vocadlo, D, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2014-04-30 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N-Acetylgalactosaminyltransferase 2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CR2
 
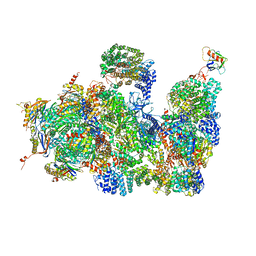 | | Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASE REGULATORY SUBUNIT 6A, 26S PROTEASE REGULATORY SUBUNIT 6B HOMOLOG, ... | | Authors: | Unverdorben, P, Beck, F, Sledz, P, Schweitzer, A, Pfeifer, G, Plitzko, J.M, Baumeister, W, Foerster, F. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-02 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Deep Classification of a Large Cryo-Em Dataset Defines the Conformational Landscape of the 26S Proteasome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6FD2
 
 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
6F62
 
 | |
