1Q6J
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 2 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, [4-(2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL](DIFLUORO)METHYLPHOSPHONIC ACID | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6M
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 3 | | Descriptor: | Protein-tyrosine phosphatase, non-receptor type 1, {[2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-(3,4-DIFLUOROPHENYL)PROPANE-1,3-DIYL]BIS[4,1-PHENYLENE(DIFLUOROMETHYLENE)]}BIS(PHOSPHONIC ACID) | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6P
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 4'-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)-1,1'-BIPHENYL-3-YLPHOSPHONIC ACID, CHLORIDE ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
1Q6S
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 9 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-((2R)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-METHYLQUINOLIN-8-YLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1OQY
 
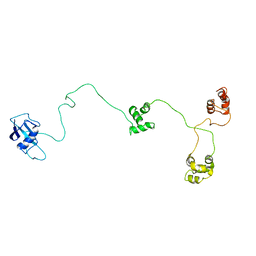 | | Structure of the DNA repair protein hHR23a | | Descriptor: | UV excision repair protein RAD23 homolog A | | Authors: | Walters, K.J, Lech, P.J, Goh, A.M, Wang, Q, Howley, P.M. | | Deposit date: | 2003-03-11 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6M1V
 
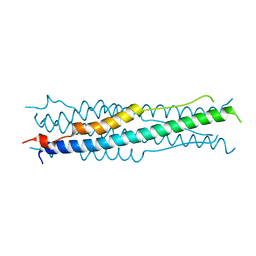 | |
4XUF
 
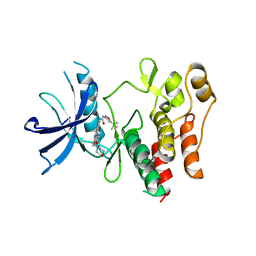 | | Crystal structure of the FLT3 kinase domain bound to the inhibitor quizartinib (AC220) | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zorn, J.A, Wang, Q, Fujimura, E, Barros, T, Kuriyan, J. | | Deposit date: | 2015-01-25 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the FLT3 Kinase Domain Bound to the Inhibitor Quizartinib (AC220).
Plos One, 10, 2015
|
|
4WJA
 
 | | Crystal Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
4IGO
 
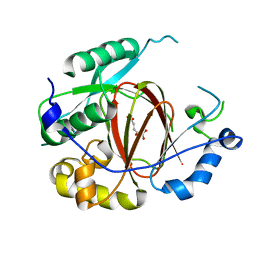 | | Histone H3 Lysine 4 Demethylating rice Rice JMJ703 in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGQ
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 in complex with methylated H3K4 substrate | | Descriptor: | FE (III) ION, N-OXALYLGLYCINE, Os05g0196500 protein, ... | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGP
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 apo enzyme | | Descriptor: | FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4J0I
 
 | | Tannin acyl hydrolase in complex with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0C
 
 | | tannin acyl hydrolase from Lactobacillus plantarum (native structure) | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Tannase | | Authors: | Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0K
 
 | | Tannin acyl hydrolase in complex with ethyl gallate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
5WT9
 
 | | Complex structure of PD-1 and nivolumab-Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Nivolumab, Light Chain of Nivolumab, ... | | Authors: | Tan, S, Zhang, H, Chai, Y, Song, H, Tong, Z, Wang, Q, Qi, J, Wong, G, Zhu, X, Liu, W.J, Gao, S, Wang, Z, Shi, Y, Yang, F, Gao, G.F, Yan, J. | | Deposit date: | 2016-12-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Nat Commun, 8, 2017
|
|
4J0J
 
 | | Tannin acyl hydrolase in complex with ethyl 3,5-dihydroxybenzoate | | Descriptor: | Tannase, ethyl 3,5-dihydroxybenzoate | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0G
 
 | | Tannin acyl hydrolase (mercury derivative) | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERCURY (II) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0H
 
 | | Tannin acyl hydrolase in complex with gallic acid | | Descriptor: | 3,4,5-trihydroxybenzoic acid, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4J0D
 
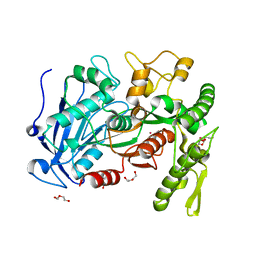 | | tannin acyl hydrolase from Lactobacillus plantarum (Cadmium) | | Descriptor: | CADMIUM ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, Chen, Q, McKinstry, W.J. | | Deposit date: | 2013-01-30 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
4JUI
 
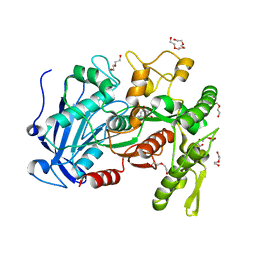 | | crystal structure of tannase from from Lactobacillus plantarum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Ren, B, Wu, M, Wang, Q, Peng, X, Wen, H, McKinstry, W.J, Chen, Q. | | Deposit date: | 2013-03-24 | | Release date: | 2013-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of tannase from Lactobacillus plantarum.
J.Mol.Biol., 425, 2013
|
|
3LUT
 
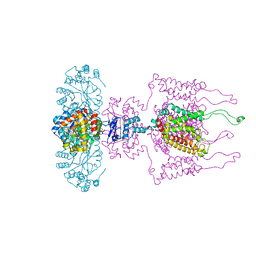 | | A Structural Model for the Full-length Shaker Potassium Channel Kv1.2 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Chen, X, Ni, F, Wang, Q, Ma, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the full-length Shaker potassium channel Kv1.2 by normal-mode-based X-ray crystallographic refinement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2PFD
 
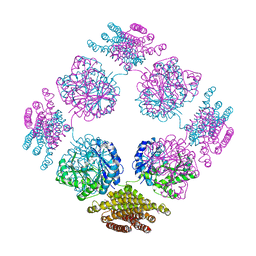 | | Anisotropically refined structure of FTCD | | Descriptor: | Formimidoyltransferase-cyclodeaminase | | Authors: | Poon, B.K, Chen, X, Lu, M, Quiocho, F.A, Wang, Q, Ma, J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Anisotropically refined structure of FTCD
To be Published
|
|
8WAN
 
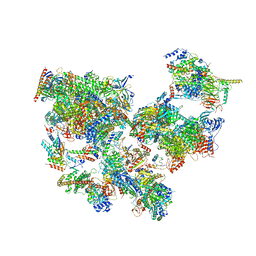 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAV
 
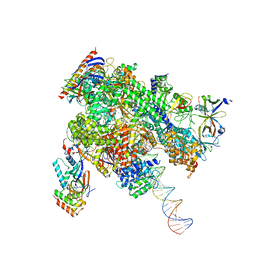 | | De novo transcribing complex 12 (TC12), the early elongation complex with Pol II positioned 12nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
