5KZC
 
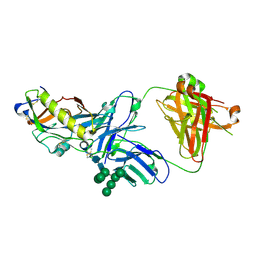 | | Crystal structure of an HIV-1 gp120 engineered outer domain with a Man9 glycan at position N276, in complex with broadly neutralizing antibody VRC01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Engineered outer domain of gp120, VRC01 Fab heavy chain, ... | | Authors: | Julien, J.-P, Jardine, J.G, Diwanji, D, Schief, W.R, Wilson, I.A. | | Deposit date: | 2016-07-24 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design.
Plos Pathog., 12, 2016
|
|
3UX8
 
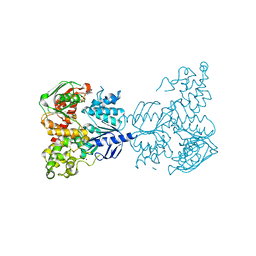 | | Crystal structure of UvrA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC, A subunit, ... | | Authors: | Samuels, M.A, Pakotiprapha, D, Jeruzalmi, D. | | Deposit date: | 2011-12-04 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of the UvrA-UvrB DNA damage sensor.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4UEC
 
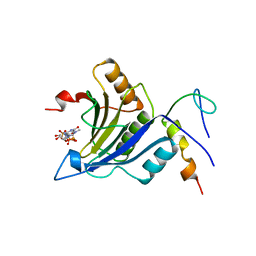 | | Complex of D. melanogaster eIF4E with eIF4G and cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4G, ... | | Authors: | Peter, D, Weichenrieder, O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Architecture of 4E-BP Translational Inhibitors Bound to Eif4E.
Mol.Cell, 57, 2015
|
|
5KWD
 
 | |
4UEA
 
 | |
5LAW
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 14 | | Descriptor: | 2-[(3~{S},3'~{a}~{S},6'~{S},6'~{a}~{S})-6-chloranyl-6'-(3-chlorophenyl)-4'-(cyclopropylmethyl)-2-oxidanylidene-spiro[1~{H}-indole-3,5'-3,3~{a},6,6~{a}-tetrahydro-2~{H}-pyrrolo[3,2-b]pyrrole]-1'-yl]ethanoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAZ
 
 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND BI-0252 | | Descriptor: | 4-[(2~{R},3~{a}~{S},5~{S},6~{S},6~{a}~{S})-6'-chloranyl-6-(3-chloranyl-2-fluoranyl-phenyl)-4-(cyclopropylmethyl)-2'-oxidanylidene-spiro[1,2,3,3~{a},6,6~{a}-hexahydropyrrolo[3,2-b]pyrrole-5,3'-1~{H}-indole]-2-yl]benzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
4UE8
 
 | |
5LAV
 
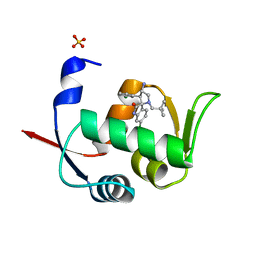 | | Novel Spiro[3H-indole-3,2 -pyrrolidin]-2(1H)-one Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) in complex with compound 6b | | Descriptor: | (3~{S},3'~{S},4'~{S})-4'-azanyl-6-chloranyl-3'-(3-chlorophenyl)-1'-(2,2-dimethylpropyl)spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
5LAY
 
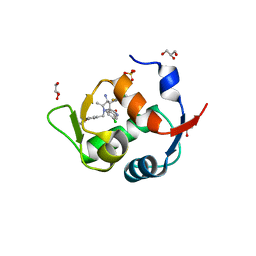 | | Discovery of New Natural-product-inspired Spiro-oxindole Compounds as Orally Active Inhibitors of the MDM2-p53 Interaction: HDM2 (MDM2) IN COMPLEX WITH COMPOUND 6g | | Descriptor: | (3~{S},3'~{S},4'~{S},5'~{S})-4'-azanyl-6-chloranyl-3'-(3-chloranyl-2-fluoranyl-phenyl)-1'-[(3-ethoxyphenyl)methyl]-5'-methyl-spiro[1~{H}-indole-3,2'-pyrrolidine]-2-one, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Kessler, D, Gollner, A. | | Deposit date: | 2016-06-15 | | Release date: | 2016-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Discovery of Novel Spiro[3H-indole-3,2'-pyrrolidin]-2(1H)-one Compounds as Chemically Stable and Orally Active Inhibitors of the MDM2-p53 Interaction.
J. Med. Chem., 59, 2016
|
|
3LPE
 
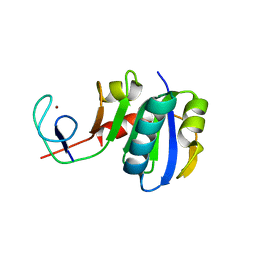 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | Descriptor: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | Authors: | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
3LIP
 
 | |
1IE9
 
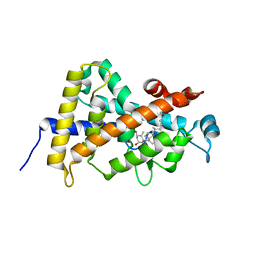 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5UJL
 
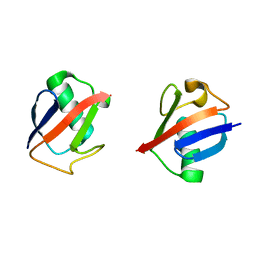 | |
1IHQ
 
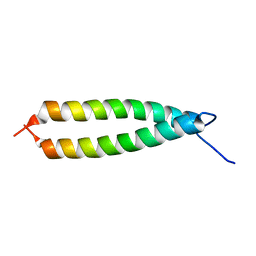 | | GLYTM1BZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF A RAT SHORT ALPHA TROPOMYOSIN WITH THE N-TERMINUS ENCODED BY EXON 1B | | Descriptor: | CHIMERIC PEPTIDE GlyTM1bZip: TROPOMYOSIN ALPHA CHAIN, BRAIN-3 and GENERAL CONTROL PROTEIN GCN4 | | Authors: | Greenfield, N.J, Yuang, Y.J, Palm, T, Swapna, G.V, Monleon, D, Montelione, G.T, Hitchcock-Degregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and folding dynamics of the N terminus of a rat non-muscle alpha-tropomyosin in an engineered chimeric protein.
J.Mol.Biol., 312, 2001
|
|
1ILX
 
 | | Excited State Dynamics in Photosystem II Revised. New Insights from the X-ray Structure. | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, CADMIUM ION, CHLOROPHYLL A, ... | | Authors: | Vasilev, S, Orth, P, Zouni, A, Owens, T.G, Bruce, D. | | Deposit date: | 2001-05-09 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Excited-state dynamics in photosystem II: insights from the x-ray crystal structure.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ISR
 
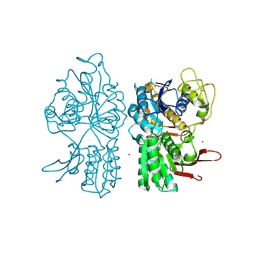 | | Crystal Structure of Metabotropic Glutamate Receptor Subtype 1 Complexed with Glutamate and Gadolinium Ion | | Descriptor: | GADOLINIUM ATOM, GLUTAMIC ACID, Metabotropic Glutamate Receptor subtype 1 | | Authors: | Tsuchiya, D, Kunishima, N, Kamiya, N, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-21 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | Descriptor: | TRF1 | | Authors: | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
1IXZ
 
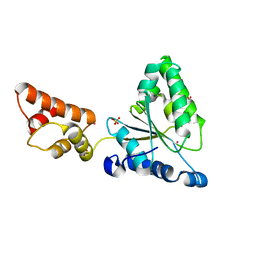 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, MERCURY (II) ION, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IO2
 
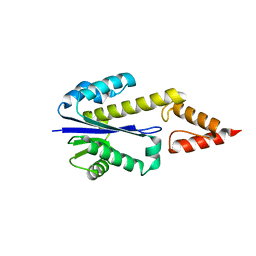 | | Crystal structure of type 2 ribonuclease h from hyperthermophilic archaeon, thermococcus kodakaraensis kod1 | | Descriptor: | RIBONUCLEASE HII | | Authors: | Muroya, A, Tsuchiya, D, Ishikawa, M, Haruki, M, Morikawa, M. | | Deposit date: | 2000-12-28 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic center of an archaeal type 2 ribonuclease H as revealed by X-ray crystallographic and mutational analyses.
Protein Sci., 10, 2001
|
|
9FYP
 
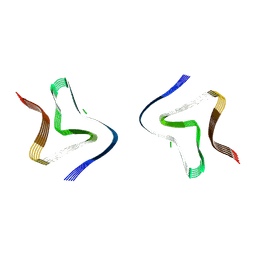 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation.
Elife, 12, 2024
|
|
5E0Q
 
 | |
1IY1
 
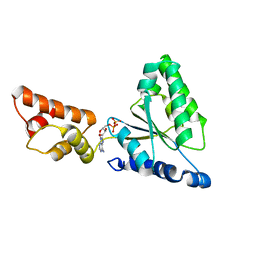 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IIZ
 
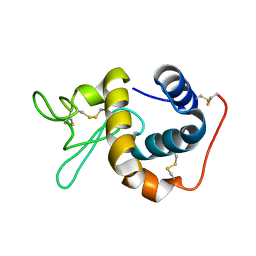 | | Crystal Structure of the Induced Antibacterial Protein from Tasar Silkworm, Antheraea mylitta | | Descriptor: | LYSOZYME | | Authors: | Jain, D, Nair, D.T, Swaminathan, G.J, Abraham, E.G, Nagaraju, J, Salunke, D.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the induced antibacterial protein from tasar silkworm, Antheraea mylitta. Implications to molecular evolution.
J.Biol.Chem., 276, 2001
|
|
1IE8
 
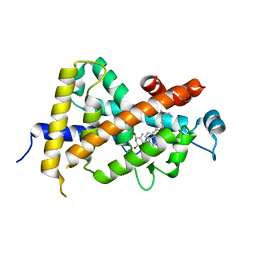 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
