7FJC
 
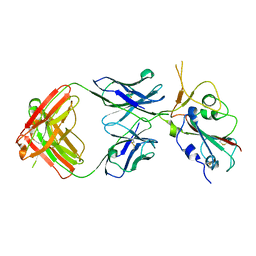 | | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab | | Descriptor: | P36-5D2 heavy chain, P36-5D2 light chain, Spike protein S1, ... | | Authors: | Zhang, L.Q, Wang, X.Q, Shan, S.S, Lan, J. | | Deposit date: | 2021-08-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of SARS-CoV-2 Beta RBD complexed with P36-5D2 Fab
To Be Published
|
|
3JAU
 
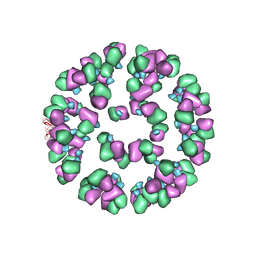 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
8WT0
 
 | | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase | | Descriptor: | Cell filamentation protein Fic, D(-)-TARTARIC ACID, DUF2559 domain-containing protein, ... | | Authors: | Chen, F.R, Guo, L.W, Lu, C.H, Jiang, W.J, Luo, Z.Q, Liu, J.F, Zhang, L.Q. | | Deposit date: | 2023-10-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The toxin-antitoxin complex Fic-1-AntF is a deAMPylase that regulates the activity of DNA gyrase
To Be Published
|
|
6JFI
 
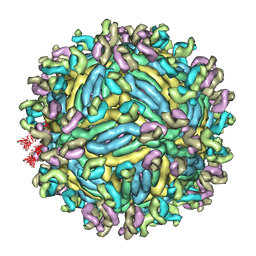 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
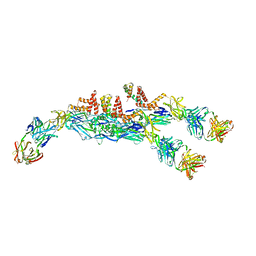 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | Descriptor: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JEP
 
 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | Descriptor: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | Authors: | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
4G8B
 
 | |
4G9E
 
 | |
4G9G
 
 | |
4G5X
 
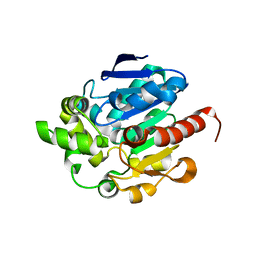 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
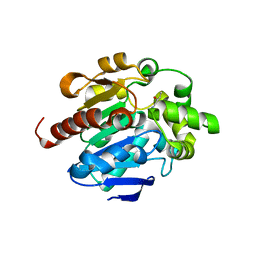
4G8C
 
 | |
4G8D
 
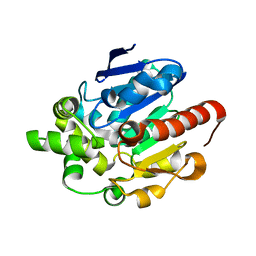 | |
4HWV
 
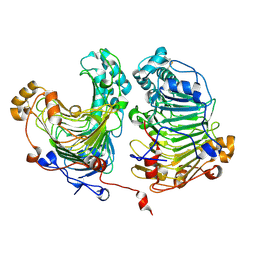 | |
6ZS1
 
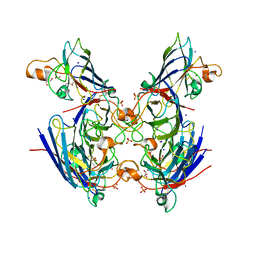 | | Chaetomium thermophilum CuZn-superoxide dismutase | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Papageorgiou, A.C, Mohsin, I. | | Deposit date: | 2020-07-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of a Cu,Zn Superoxide Dismutase From the Thermophilic Fungus Chaetomium thermophilum.
Protein Pept.Lett., 28, 2021
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | Descriptor: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2K
 
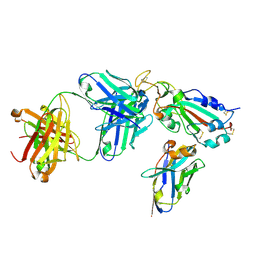 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | Descriptor: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7E1N
 
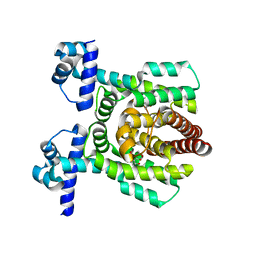 | | Crystal structure of PhlH in complex with 2,4-diacetylphloroglucinol | | Descriptor: | 2,4-bis[(1R)-1-oxidanylethyl]benzene-1,3,5-triol, DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-02 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E1L
 
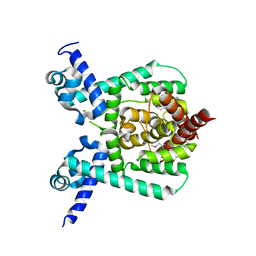 | | Crystal structure of apo form PhlH | | Descriptor: | DUF1956 domain-containing protein | | Authors: | Zhang, N, Wu, J, He, Y.X, Ge, H. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-02 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for coordinating secondary metabolite production by bacterial and plant signaling molecules.
J.Biol.Chem., 298, 2022
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
7D0C
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3A1, ... | | Authors: | Yan, R.H, Wang, R.K, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0B
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_1B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
7D0D
 
 | | S protein of SARS-CoV-2 in complex bound with P5A-3C12_2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of P5A-3C12, ... | | Authors: | Yan, R.H, Wang, R.K, Ju, B, Yu, J.F, Zhang, Y.Y, Liu, N, Wang, H.W, Wang, X.Q, Zhang, L.Q, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
