1I84
 
 | | CRYO-EM STRUCTURE OF THE HEAVY MEROMYOSIN SUBFRAGMENT OF CHICKEN GIZZARD SMOOTH MUSCLE MYOSIN WITH REGULATORY LIGHT CHAIN IN THE DEPHOSPHORYLATED STATE. ONLY C ALPHAS PROVIDED FOR REGULATORY LIGHT CHAIN. ONLY BACKBONE ATOMS PROVIDED FOR S2 FRAGMENT. | | Descriptor: | SMOOTH MUSCLE MYOSIN ESSENTIAL LIGHT CHAIN, SMOOTH MUSCLE MYOSIN HEAVY CHAIN, SMOOTH MUSCLE MYOSIN REGULATORY LIGHT CHAIN | | Authors: | Wendt, T, Taylor, D, Trybus, K.M, Taylor, K. | | Deposit date: | 2001-03-12 | | Release date: | 2001-03-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON CRYSTALLOGRAPHY (20 Å) | | Cite: | Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3J5Y
 
 | | Structure of the mammalian ribosomal pre-termination complex associated with eRF1-eRF3-GDPNP | | Descriptor: | 5'-R(*AP*UP*UP*GP*UP*AP*AP*AP*AP*A)-3', Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, Eukaryotic peptide chain release factor subunit 1, ... | | Authors: | des Georges, A, Hashem, Y, Unbehaun, A, Grassucci, R.A, Taylor, D, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2013-11-21 | | Release date: | 2013-12-25 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structure of the mammalian ribosomal pre-termination complex associated with eRF1*eRF3*GDPNP.
Nucleic Acids Res., 42, 2014
|
|
7L7H
 
 | | Alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Hojjatian, A, Dasari, A. | | Deposit date: | 2020-12-28 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Distinct cryo-EM Structure of Alpha-synuclein Filaments derived by Tau
To Be Published
|
|
6NTS
 
 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
3J04
 
 | | EM structure of the heavy meromyosin subfragment of Chick smooth muscle Myosin with regulatory light chain in phosphorylated state | | Descriptor: | Myosin light polypeptide 6, Myosin regulatory light chain 2, smooth muscle major isoform, ... | | Authors: | Baumann, B.A.J, Taylor, D, Huang, Z, Tama, F, Fagnant, P.M, Trybus, K, Taylor, K. | | Deposit date: | 2011-02-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation.
J.Mol.Biol., 415, 2012
|
|
7N9T
 
 | |
3DTP
 
 | | Tarantula heavy meromyosin obtained by flexible docking to Tarantula muscle thick filament Cryo-EM 3D-MAP | | Descriptor: | Myosin II regulatory light chain, Myosin light polypeptide 6, Myosin-11,Myosin-7 | | Authors: | Alamo, L, Wriggers, W, Pinto, A, Bartoli, F, Salazar, L, Zhao, F.Q, Craig, R, Padron, R. | | Deposit date: | 2008-07-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Three-Dimensional Reconstruction of Tarantula Myosin Filaments Suggests How Phosphorylation May Regulate Myosin Activity
J.Mol.Biol., 384, 2008
|
|
8FD8
 
 | | human 15-PGDH with NADH bound | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)] | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-12-02 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
3JAX
 
 | | Heavy meromyosin from Schistosoma mansoni muscle thick filament by negative stain EM | | Descriptor: | myosin 2 heavy chain, myosin regulatory light chain, smooth muscle myosin essential light chain | | Authors: | Sulbaran, G, Alamo, L, Pinto, A, Marquez, G, Mendez, F, Padron, R, Craig, R. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | An invertebrate smooth muscle with striated muscle myosin filaments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2XKF
 
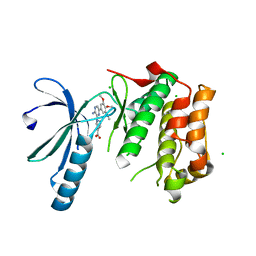 | | Structure of Nek2 bound to aminopyrazine compound 2 | | Descriptor: | 1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKD
 
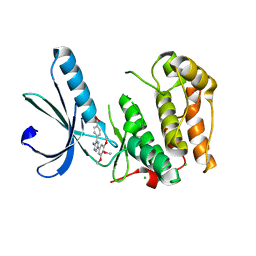 | | Structure of Nek2 bound to aminopyrazine compound 12 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]benzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK3
 
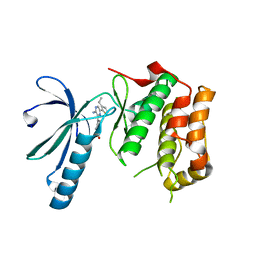 | | Structure of Nek2 bound to Aminopyrazine compound 35 | | Descriptor: | 4-[3-AMINO-6-(3-ETHYLTHIOPHEN-2-YL)PYRAZIN-2-YL]CYCLOHEXANE-1-CARBOXYLIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK4
 
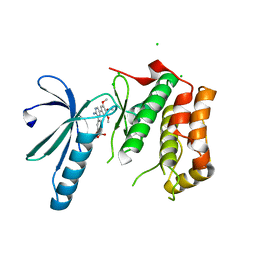 | | Structure of Nek2 bound to aminopyrazine compound 17 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-ethoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK8
 
 | | Structure of Nek2 bound to aminopyrazine compound 15 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-methoxybenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK7
 
 | | Structure of Nek2 bound to aminopyrazine compound 23 | | Descriptor: | (3R,4R)-1-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-3-ethylpiperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKE
 
 | | Structure of Nek2 bound to Aminipyrazine Compound 5 | | Descriptor: | 1-[3-amino-6-(3-methoxyphenyl)pyrazin-2-yl]piperidine-4-carboxylic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XKC
 
 | | Structure of Nek2 bound to aminopyrazine compound 14 | | Descriptor: | 4-[3-amino-6-(3,4,5-trimethoxyphenyl)pyrazin-2-yl]-2-methylbenzoic acid, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
2XK6
 
 | | Structure of Nek2 bound to aminopyrazine compound 36 | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2, cis-4-[3-amino-6-(3-cyclopropylthiophen-2-yl)pyrazin-2-yl]cyclohexanecarboxylic acid | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2010-07-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aminopyrazine Inhibitors Binding to an Unusual Inactive Conformation of the Mitotic Kinase Nek2: Sar and Structural Characterization.
J.Med.Chem., 53, 2010
|
|
5FB8
 
 | | Structure of Interleukin-16 bound to the 14.1 antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Anti-IL-16 antibody 14.1 Fab domain Heavy Chain, ... | | Authors: | Hall, G, Cowan, R, Bayliss, R, Carr, M. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a Potential Therapeutic Antibody Bound to Interleukin-16 (IL-16): MECHANISTIC INSIGHTS AND NEW THERAPEUTIC OPPORTUNITIES.
J.Biol.Chem., 291, 2016
|
|
7KHA
 
 | | Cryo-EM Structure of the Desulfovibrio vulgaris Type I-C Apo Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, CT1134 family, ... | | Authors: | O'Brien, R, Wrapp, D, Bravo, J.P.K, Schwartz, E, Taylor, D. | | Deposit date: | 2020-10-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for assembly of non-canonical small subunits into type I-C Cascade.
Nat Commun, 11, 2020
|
|
7SMU
 
 | | Crystal Structure of Consomatin-Ro1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45-pentadecaoxaoctatetracontane-1,48-diol, Consomatin-Ro1 | | Authors: | Ramiro, I.B.L, Whitby, F.G, Hill, C.P, Safavi-Hemami, H, Concepcion, G.P, Olivera, B.M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Somatostatin venom analogs evolved by fish-hunting cone snails: From prey capture behavior to identifying drug leads.
Sci Adv, 8, 2022
|
|
7KOG
 
 | | Lethocerus Myosin II complete coiled-coil domain resolved in its native environment | | Descriptor: | Myosin heavy chain isoform Mhc_X1 | | Authors: | Rahmani, H, Hu, Z, Daneshparvar, N, Taylor, D, Taylor, K.A. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-24 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The myosin II coiled-coil domain atomic structure in its native environment.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4DCQ
 
 | | Crystal Structure of the Fab Fragment of 3B5H10, an Antibody-Specific for Extended Polyglutamine Repeats (orthorhombic form) | | Descriptor: | 1,2-ETHANEDIOL, 3B5H10 FAB Heavy Chain, 3B5H10 FAB Light Chain | | Authors: | Peters-Libeu, C.A, Tran, T, Finkbeiner, S, Weisgraber, K. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
3S96
 
 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
8CYA
 
 | | SARS-CoV-2 Spike protein in complex with a pan-sarbecovirus nanobody 2-67 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
