2KZR
 
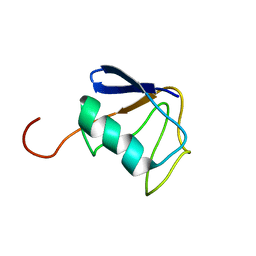 | | Solution NMR Structure of Ubiquitin thioesterase OTU1 (EC 3.1.2.-) from Mus musculus, Northeast Structural Genomics Consortium Target MmT2A | | Descriptor: | Ubiquitin thioesterase OTU1 | | Authors: | Chitayat, S, Gutmanas, A, Lemak, A, Yee, A, Bezsonova, I, Wu, B, Doherty, R.S, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Dhe-Paganon, S, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-23 | | Release date: | 2010-07-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target MmT2A
To be Published
|
|
2O4E
 
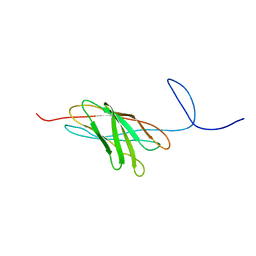 | | The solution structure of a protein-protein interaction module from a family 84 glycoside hydrolase of Clostridium perfringens | | Descriptor: | O-GlcNAcase nagJ | | Authors: | Chitayat, S, Adams, J.J, Gregg, K, Boraston, A.B, Smith, S.P. | | Deposit date: | 2006-12-04 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a putative non-cellulosomal cohesin module from a Clostridium perfringens family 84 glycoside hydrolase.
J.Mol.Biol., 375, 2008
|
|
2JH2
 
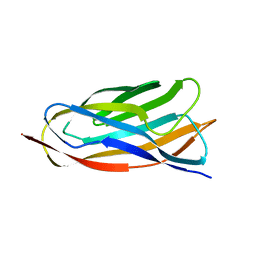 | | X-ray crystal structure of a cohesin-like module from Clostridium perfringens | | Descriptor: | O-GLCNACASE NAGJ | | Authors: | Chitayat, S, Gregg, K, Adams, J.J, Ficko-Blean, E, Bayer, E.A, Boraston, A.B, Smith, S.P. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-Dimensional Structure of a Putative Non- Cellulosomal Cohesin Module from a Clostridium Perfringens Family 84 Glycoside Hydrolase.
J.Mol.Biol., 375, 2008
|
|
2FEB
 
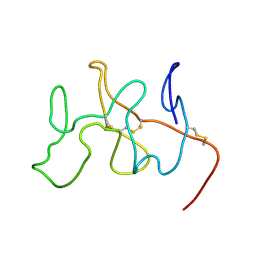 | | NMR Solution Structure, Dynamics and Binding Properties of the Kringle IV Type 8 module of apolipoprotein(a) | | Descriptor: | Apolipoprotein(a) | | Authors: | Chitayat, S, Kanelis, V, Koschinsky, M.L, Smith, S.P. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance (NMR) solution structure, dynamics, and binding properties of the kringle IV type 8 module of apolipoprotein(a).
Biochemistry, 46, 2007
|
|
2JNK
 
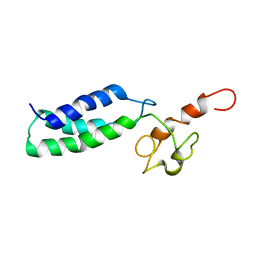 | |
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
5TLQ
 
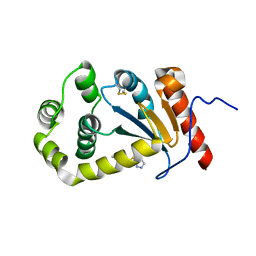 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
1ISH
 
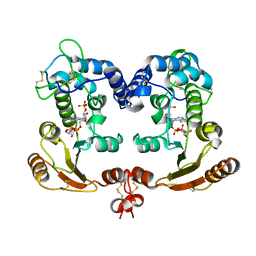 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHENO-NADP, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISF
 
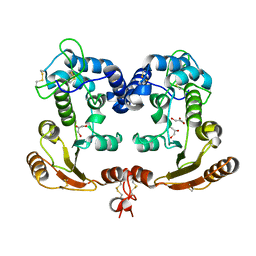 | | Crystal Structure Analysis of BST-1/CD157 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISJ
 
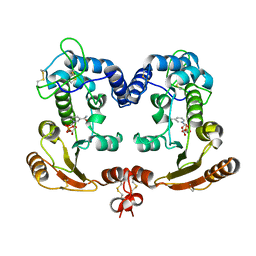 | | Crystal Structure Analysis of BST-1/CD157 complexed with NMN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISI
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, [[(2R,3S,4R,5R)-3,4-dihydroxy-5-(9H-imidazo[2,1-f]purin-6-ium-3-yl)oxolan-2-yl]methoxy-oxidanidyl-phosphoryl] [(2R,3S,4R,5R)-3,4-dihydroxy-5-oxidanidyl-oxolan-2-yl]methyl phosphate, ... | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISG
 
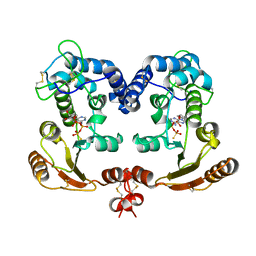 | | Crystal Structure Analysis of BST-1/CD157 with ATPgammaS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISM
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with nicotinamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
5VOE
 
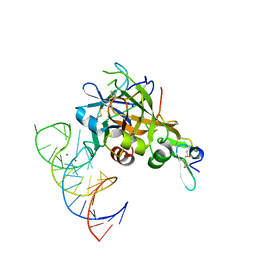 | | DesGla-XaS195A Bound to Aptamer 11F7t | | Descriptor: | Aptamer 11F7t (36-MER), CALCIUM ION, Coagulation factor X, ... | | Authors: | Gunaratne, R, Kumar, S, Frederiksen, J.W, Stayrook, S, Lohrmann, J.L, Perry, K, Chabata, C.V, Thalji, N.K, Ho, M.D, Arepally, G, Camire, R.M, Krishnaswamy, S.K, Sullenger, B.A. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
2LFE
 
 | | Solution NMR structure of N-terminal domain of human E3 ubiquitin-protein ligase HECW2, Northeast structural genomics consortium (NESG) target ht6306A | | Descriptor: | E3 ubiquitin-protein ligase HECW2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Chitayat, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of E3 ligase HECW2
To be Published
|
|
2KWF
 
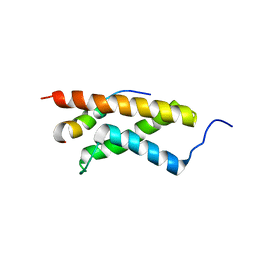 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
2LS6
 
 | | Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringens | | Descriptor: | Hyaluronoglucosaminidase | | Authors: | Grondin, J.M, Chitayat, S, Ficko-Blean, E, Boraston, A.B, Smith, S.P. | | Deposit date: | 2012-04-20 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An unusual mode of galactose recognition by a family 32 carbohydrate-binding module.
J.Mol.Biol., 426, 2014
|
|
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
2M1U
 
 | | Solution structure of the small dictyostelium discoideium myosin light chain mlcb provides insights into iq-motif recognition of class i myosin myo1b | | Descriptor: | myosin light chain MlcB | | Authors: | Liburd, J, Chitayat, S, Crawley, S.W, Denis, C.M, Cote, G.P, Smith, S.P. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the small Dictyostelium discoideum myosin light chain MlcB provides insights into MyoB IQ motif recognition.
J.Biol.Chem., 289, 2014
|
|
2M8U
 
 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | Descriptor: | Myosin Light Chain, MlcC | | Authors: | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | Deposit date: | 2013-05-28 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2MH0
 
 | |
4O21
 
 | | Product complex of metal-free PKAc, ATP-gamma-S and SP20. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thio-phosphorylated peptide pSP20, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Das, A, Kovalevsky, A.Y, Gerlits, O, Langan, P, Heller, W.T, Keshwani, M, Taylor, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metal-Free cAMP-Dependent Protein Kinase Can Catalyze Phosphoryl Transfer.
Biochemistry, 53, 2014
|
|
2YM4
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1, ethyl 4-[(2R)-2-(aminomethyl)morpholin-4-yl]-3-(3-cyanophenyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxylate | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YM6
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2R)-4-(9H-pyrido[4',3':4,5]pyrrolo[2,3-d]pyrimidin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
