7VBG
 
 | | N Terminal Domain of PRC1 | | Descriptor: | Protein regulator of cytokinesis 1 | | Authors: | Tan, F, Xia, B. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | N Terminal Domain of PRC1
To Be Published
|
|
4ZD6
 
 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4Z9F
 
 | | Halohydrin hydrogen-halide-lyase, HheA | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase A | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-10 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
4ZU3
 
 | | Halohydrin hydrogen-halide-lyases, HheB | | Descriptor: | 3-hydroxypentanedinitrile, Halohydrin epoxidase B, MAGNESIUM ION, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-05-15 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
5B12
 
 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
8C7F
 
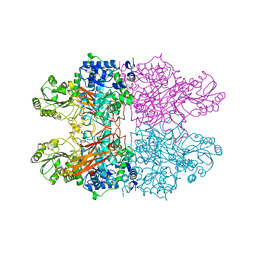 | |
8RDU
 
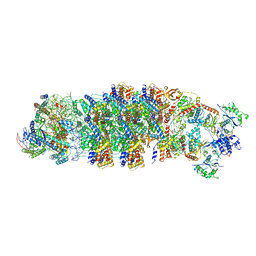 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LE, MAGNESIUM ION, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKU
 
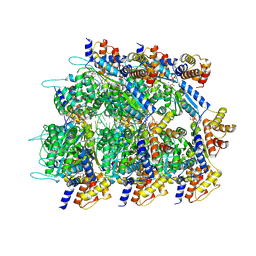 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKT
 
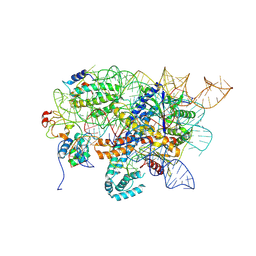 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K Cas12k domain local-refinement map | | Descriptor: | MAGNESIUM ION, Non-target strand - LE, ShCas12k, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8RKV
 
 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsB domain local-refinement map | | Descriptor: | LE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
7B7O
 
 | | Solution structure of A. thaliana core TatA in DHPC micelles | | Descriptor: | Sec-independent protein translocase protein TATA, chloroplastic | | Authors: | Pettersson, P, Ye, W, Jakob, M, Tannert, F, Klosgen, R.B, Maler, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR.
FEBS J, 285, 2018
|
|
2NSM
 
 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
7P65
 
 | | Progressive supranuclear palsy tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P66
 
 | | Globular glial tauopathy type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P68
 
 | | Globular glial tauopathy type 3 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P67
 
 | | Globular glial tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6C
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6E
 
 | | Argyrophilic grain disease type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6D
 
 | | Argyrophilic grain disease type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6A
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1a tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6B
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 1b tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
2DQU
 
 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DQT
 
 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
7PY2
 
 | | Structure of pathological TDP-43 filaments from ALS with FTLD | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Arseni, D, Hasegawa, H, Murzin, A.G, Kametani, F, Arai, M, Yoshida, M, Falcon, B. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure of pathological TDP-43 filaments from ALS with FTLD.
Nature, 601, 2022
|
|
8AA5
 
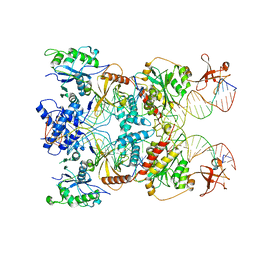 | | Cryo-EM structure of the strand transfer complex of the TnsB transposase (type V-K CRISPR-associated transposon) | | Descriptor: | LE_PolyA, LE_Target, RE_PolyA, ... | | Authors: | Tenjo-Castano, F, Sofos, N, Lopez-Mendez, B, Stutzke, L.S, Fuglsang, A, Stella, S, Montoya, G. | | Deposit date: | 2022-06-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structure of the TnsB transposase-DNA complex of type V-K CRISPR-associated transposon.
Nat Commun, 13, 2022
|
|
