6K16
 
 | |
8H8X
 
 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8HAE
 
 | | Cryo-EM structure of HACE1 dimer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
1KFT
 
 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
2GKD
 
 | |
2L65
 
 | | HADDOCK calculated model of the complex of the resistance protein CalC and Calicheamicin-Gamma | | Descriptor: | 2,4-dideoxy-4-(ethylamino)-3-O-methyl-alpha-L-threo-pentopyranose-(1-2)-4-amino-4,6-dideoxy-beta-D-glucopyranose, 2,6-dideoxy-4-thio-beta-D-allopyranose, 3-O-methyl-alpha-L-rhamnopyranose, ... | | Authors: | Singh, S, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
8CMU
 
 | | High resolution structure of the coagulation Factor XIII A2B2 heterotetramer complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Urgular, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | The cryo-EM structure of the plasma coagulation Factor XIII complex at 2.68 Angstrom resolution.
To Be Published
|
|
8CMT
 
 | | Structure of the plasma coagulation Factor XIII A2B2 heterotetrameric complex. | | Descriptor: | Coagulation factor XIII A chain, Coagulation factor XIII B chain | | Authors: | Singh, S, Ugurlar, D, Hagelueken, G, Geyer, M, Biswas, A. | | Deposit date: | 2023-02-21 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | The cryo-EM structure of plasma coagulation Factor XIII heterotetrameric complex at 2.68 Angstrom resolution.
To Be Published
|
|
6MRO
 
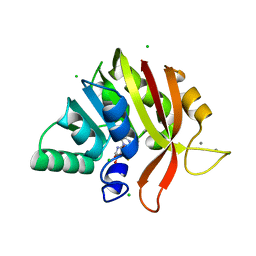 | | Crystal structure of methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution, Northeast Structural Genomics Consortium (NESG) Target MvR53. | | Descriptor: | CALCIUM ION, CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Forouhar, F, Wang, C, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-10-15 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a methyl transferase from Methanosarcina acetivorans at 1.6 Angstroms resolution.
To Be Published
|
|
7E6W
 
 | |
7E9R
 
 | |
7MU4
 
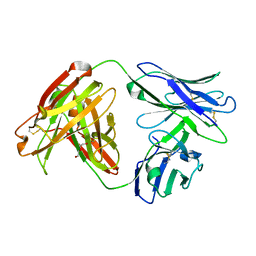 | | Crystal Structure of HPV L1-directed D24.M01Fab | | Descriptor: | D24.M01 Fab Heavy Chain, D24.M01 Fab Light Chain, DI(HYDROXYETHYL)ETHER | | Authors: | Singh, S, Pancera, M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Characterisation of Immune Responses to HPV Vaccination
To Be Published
|
|
7MX8
 
 | |
7MYT
 
 | |
3GXO
 
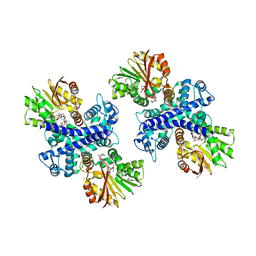 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | Descriptor: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3GWZ
 
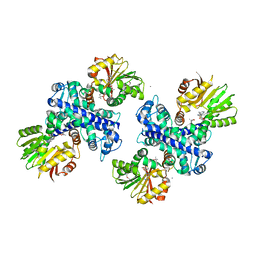 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
5YDD
 
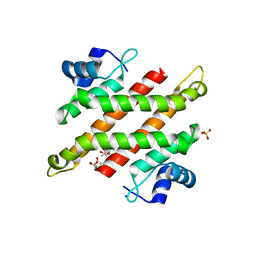 | | Crystal structure of C-terminal domain of Rv1828 from Mycobacterium tuberculosis | | Descriptor: | (6R,8S,9S)-8-(hydroxymethyl)-6,11,11-tris(oxidanyl)-9-propyl-dodecanoic acid, GLYCEROL, SODIUM ION, ... | | Authors: | Singh, S, Karthiekeyan, S. | | Deposit date: | 2017-09-12 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characteristics of the essential pathogenicity factor Rv1828, a MerR family transcription regulator from Mycobacterium tuberculosis.
FEBS J., 285, 2018
|
|
5YDC
 
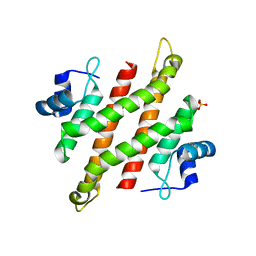 | |
1ZXF
 
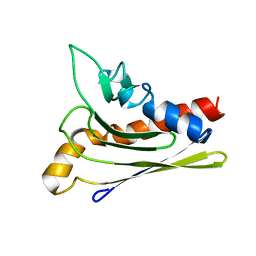 | | Solution structure of a self-sacrificing resistance protein, CalC from Micromonospora echinospora | | Descriptor: | CalC | | Authors: | Singh, S, Hager, M.H, Zhang, C, Griffith, B.R, Lee, M.S, Hallenga, K, Markley, J.L, Thorson, J.S, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-06-08 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the self-sacrifice mechanism of enediyne resistance.
Acs Chem.Biol., 1, 2006
|
|
1XO3
 
 | | Solution Structure of Ubiquitin like protein from Mus Musculus | | Descriptor: | RIKEN cDNA 2900073H19 | | Authors: | Singh, S, Tonelli, M, Tyler, R.C, Bahrami, A, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-05 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the AAH26994.1 protein from Mus musculus, a putative eukaryotic Urm1.
Protein Sci., 14, 2005
|
|
1XO8
 
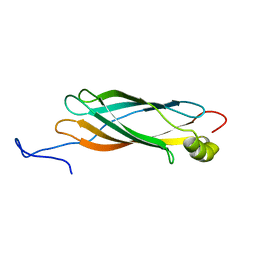 | | Solution structure of AT1g01470 from Arabidopsis Thaliana | | Descriptor: | At1g01470 | | Authors: | Singh, S, Cornilescu, C.C, Tyler, R.C, Cornilescu, G, Tonelli, M, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-06 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein
Protein Sci., 14, 2005
|
|
6K2C
 
 | | Extended Hect domain of UBE3C E3 Ligase | | Descriptor: | Ubiquitin-protein ligase E3C | | Authors: | Sivaraman, J, Singh, S. | | Deposit date: | 2019-05-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of HECT domain of UBE3C E3 ligase and its ubiquitination activity.
Biochem.J., 477, 2020
|
|
6JX6
 
 | | Tetrameric form of Smac | | Descriptor: | Diablo homolog, mitochondrial | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6JX5
 
 | | Hect domain of AREL1 | | Descriptor: | Apoptosis-resistant E3 ubiquitin protein ligase 1 | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
6WAS
 
 | | Structure of D19.PA8 Fab in complex with 1FD6 16055 V1V2 scaffold | | Descriptor: | 1FD6 16055 V1V2 scaffold, 2-acetamido-2-deoxy-beta-D-glucopyranose, GN1_PA8 Fab Heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-03-26 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
