5EQU
 
 | | Crystal structure of the epimerase SnoN in complex with Fe3+, alpha ketoglutarate and nogalamycin RO | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Nogalamycin RO, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-13 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5ERL
 
 | | Crystal structure of the epimerase SnoN in complex with Ni2+, succinate and nogalamycin RO | | Descriptor: | NICKEL (II) ION, Nogalamycin RO, SUCCINIC ACID, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-14 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EP9
 
 | | Crystal structure of the non-heme alpha ketoglutarate dependent epimerase SnoN from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, FE (III) ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Niiranen, L, Siitonen, V, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EPA
 
 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4AN4
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, GLYCOSYL TRANSFERASE | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-15 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4AMB
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4AMG
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
5BKA
 
 | |
6VW4
 
 | |
6P77
 
 | | 2.5 Angstrom structure of Caci_6494 from Catenulispora Acidiphila | | Descriptor: | Aromatic-ring-hydroxylating dioxygenase beta subunit, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Vuksanovic, N, Silvaggi, N.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural characterization of three noncanonical NTF2-like superfamily proteins: implications for polyketide biosynthesis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6P7L
 
 | |
7OWB
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-hydroxylase CalMB | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7OY1
 
 | | DnrK mutant RTCR | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,5,7-trihydroxy-6,11-dioxo-4-{[2,3,6-trideoxy-3-(dimethylamino)-alpha-L-lyxo-hexopyranosyl]oxy}-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
8R2E
 
 | |
8R2J
 
 | |
8R2B
 
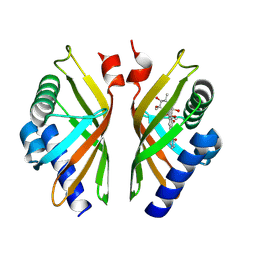 | |
8R20
 
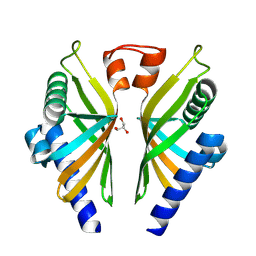 | |
7PHF
 
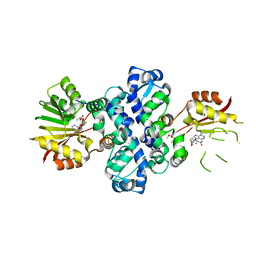 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PGA
 
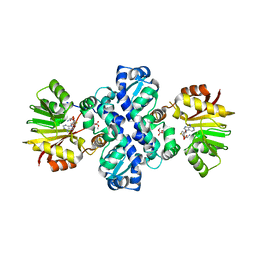 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PHD
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Grocholski, T, Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PHE
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PGJ
 
 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-decarboxylate TamK and 10-hydroxylase RdmB, together with a single point mutation F297G | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-7-methoxy-2,4,5-tris(oxidanyl)-6,11-bis(oxidanylidene)-3,4-dihydro-1H-tetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7PG7
 
 | |
