7W2I
 
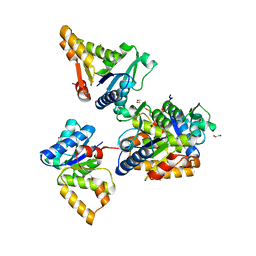 | | Crystal structure of LOG (Rv1205) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Cytokinin riboside 5'-monophosphate phosphoribohydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shang, L, Zhang, G. | | Deposit date: | 2021-11-23 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cytokinin-producing enzyme "lonely guy" (LOG) from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 598, 2022
|
|
7E0B
 
 | |
5BPE
 
 | | Crystal structure of EV71 3Cpro in complex with a potent and selective Inhibitor | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{S})-1-cyano-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-3-phenyl-2-[[(~{E})-3-phenylprop-2-enoyl]amino]propanamide, EV71 3Cpro | | Authors: | Luqing, S, Yin, Z. | | Deposit date: | 2015-05-28 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanohydrin as an Anchoring Group for Potent and Selective Inhibitors of Enterovirus 71 3C Protease
J.Med.Chem., 58, 2015
|
|
6LNQ
 
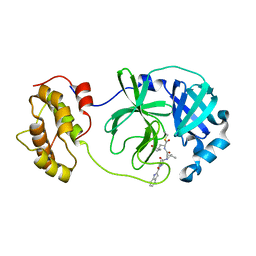 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | Descriptor: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | Authors: | Wang, H, Shang, L.Q. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
6LO0
 
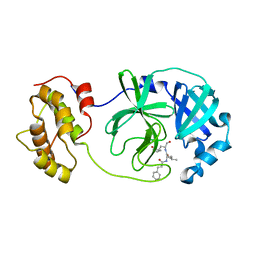 | |
6LNY
 
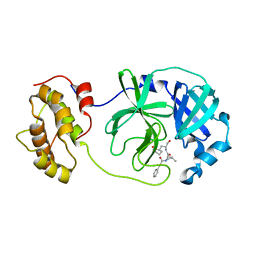 | |
6LJ9
 
 | |
6LJB
 
 | |
5GSW
 
 | | Crystal structure of EV71 3C in complex with N69S 1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-17 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
5GSO
 
 | | Crystal Structures of EV71 3C Protease in complex with NK-1.8k | | Descriptor: | 3C protein, ~{N}-[(2~{S})-3-(4-fluorophenyl)-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]propan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Wang, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Enterovirus 71 3C Protease in Complex with NK-1.8k and Indications for the Development of Antienterovirus Protease Inhibitor
Antimicrob. Agents Chemother., 61, 2017
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
7DHJ
 
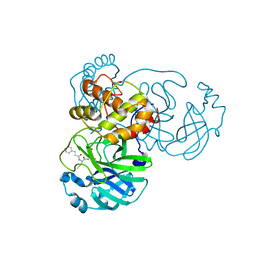 | | The co-crystal structure of SARS-CoV-2 main protease with the peptidomimetic inhibitor (S)-2-cinnamamido-N-((S)-1-oxo-3-((S)-2-oxopyrrolidin-3-yl)propan-2-yl)pent-4-ynamide | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[(~{E})-3-phenylprop-2-enoyl]amino]pent-4-ynamide, 3C-like proteinase | | Authors: | Shang, L.Q, Wang, H, Deng, W.L, Xing, S, Wang, Y.X. | | Deposit date: | 2020-11-15 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate.
Eur.J.Med.Chem., 238, 2022
|
|
3I94
 
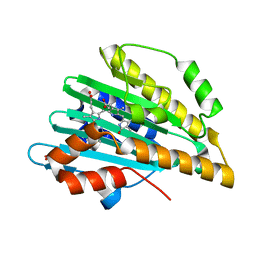 | | Crystal structure of PcyA-biliverdin XIII alpha complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase, SULFATE ION | | Authors: | Hagiwara, Y, Sugishima, M, Fukuyama, K. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural insights into vinyl reduction regiospecificity of phycocyanobilin:ferredoxin oxidoreductase (PcyA).
to be published
|
|
3I8U
 
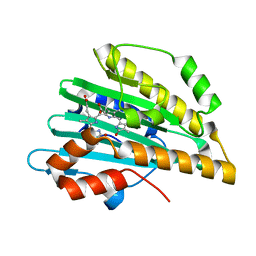 | | Crystal structure of PcyA-181,182-dihydrobiliverdin complex | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(3-ethenyl-4-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-2-ylidene]methy l]-5-[(Z)-(4-ethyl-3-methyl-5-oxo-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Phycocyanobilin:ferredoxin oxidoreductase | | Authors: | Hagiwara, Y, Sugishima, M, Fukuyama, K. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into vinyl reduction regiospecificity of phycocyanobilin:ferredoxin oxidoreductase (PcyA).
To be Published
|
|
3I95
 
 | |
7WOH
 
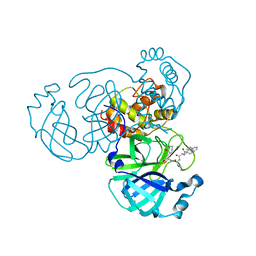 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(2S)-3-phenyl-2-[[(E)-3-phenylprop-2-enoyl]amino]propanoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO1
 
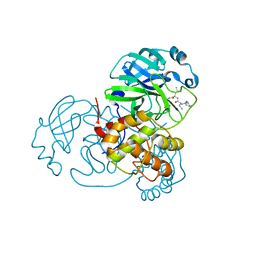 | | Discovery of SARS-CoV-2 3CLpro peptidomimetic inhibitors through H41-specific protein-ligand interactions | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]cyclohexanecarboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO3
 
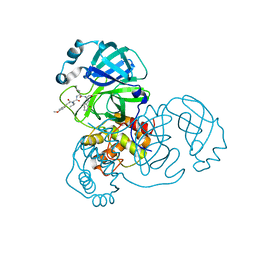 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S)-2-[[(2S)-2-[[(E)-3-(4-methoxyphenyl)prop-2-enoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WOF
 
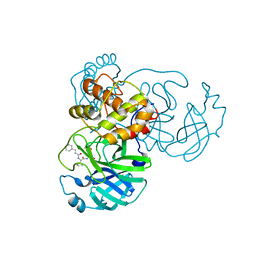 | | SARS-CoV-2 3CLpro | | Descriptor: | (2S,3S)-3-methyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-2-[[(E)-3-phenylprop-2-enoyl]amino]pentanamide, 3C-like proteinase | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
7WO2
 
 | | SARS-CoV-2 3CLPro Peptidomimetic Inhibitor TPM5 | | Descriptor: | 3C-like proteinase, N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S}-2-oxidanylidenepiperidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]furan-2-carboxamide | | Authors: | Wang, Y, Ye, S. | | Deposit date: | 2022-01-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of SARS-CoV-2 3CL Pro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions.
Int J Mol Sci, 23, 2022
|
|
