2MC7
 
 | | Structure of Salmonella MgtR | | Descriptor: | Regulatory peptide | | Authors: | Jean-Francois, F, Dai, J, Yu, L, Myrick, A, Rubin, E, Fajer, P, Song, L, Zhou, H, Cross, T. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Binding of MgtR, a Salmonella Transmembrane Regulatory Peptide, to MgtC, a Mycobacterium tuberculosis Virulence Factor: A Structural Study.
J.Mol.Biol., 426, 2014
|
|
4S1W
 
 | | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] | | Authors: | Filippova, E.V, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-01-15 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of a putative Glutamine--Fructose-6-Phosphate Aminotransferase from Staphylococcus aureus subsp. aureus Mu50
To be Published
|
|
4RTF
 
 | | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaK, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Endres, M, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-11-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of molecular chaperone DnaK from Mycobacterium tuberculosis H37Rv
To be Published
|
|
3TY7
 
 | | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, Y, Joachimiak, G, Jedrzejczak, R, Rubin, E, Ioerger, T, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Aldehyde Dehydrogenase family Protein from Staphylococcus aureus
To be Published
|
|
4JG2
 
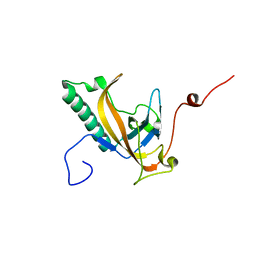 | | Structure of phage-related protein from Bacillus cereus ATCC 10987 | | Descriptor: | Phage-related protein | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of phage-related protein from Bacillus cereus ATCC 10987
To be Published
|
|
4N1X
 
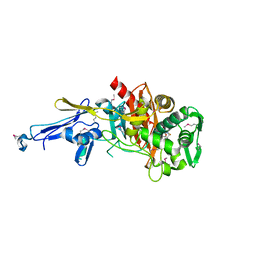 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G | | Descriptor: | OPEN FORM - PENICILLIN G, Peptidoglycan glycosyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with penicillin G
To be Published
|
|
4MQB
 
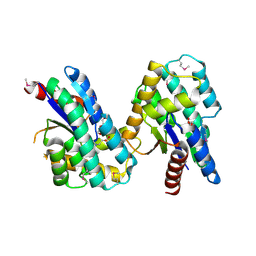 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
4JBF
 
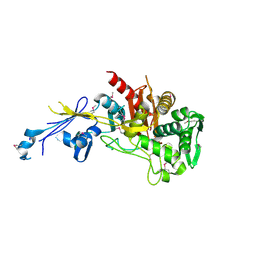 | | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469. | | Descriptor: | Peptidoglycan glycosyltransferase, TETRAETHYLENE GLYCOL | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-02-19 | | Release date: | 2013-03-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of peptidoglycan glycosyltransferase from Atopobium parvulum DSM 20469.
To be Published
|
|
4F4I
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form | | Descriptor: | Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form
To be Published
|
|
4DQ1
 
 | | Thymidylate synthase from Staphylococcus aureus. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Osipiuk, J, Holowicki, J, Jedrzejczak, R, Rubin, E, Guinn, K, Ioerger, T, Baker, D, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-14 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Thymidylate synthase from Staphylococcus aureus.
To be Published
|
|
4RPI
 
 | | Crystal Structure of Nicotinate Mononucleotide Adenylyltransferase from Mycobacterium tuberculosis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, nicotinate-nucleotide adenylyltransferase | | Authors: | Rodionova, I, Zuccola, H, Sorci, L, Aleshin, A.E, Kazanov, M, Sergienko, E, Rubin, E, Locher, C, Osterman, A. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Mycobacterial nicotinate mononucleotide adenylyltransferase: structure, mechanism, and implications for drug discovery.
J. Biol. Chem., 290, 2015
|
|
5CRF
 
 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5CXW
 
 | | Structure of the PonA1 protein from Mycobacterium Tuberculosis in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
8A8V
 
 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8W
 
 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8U
 
 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
4EAQ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with 3'-Azido-3'-Deoxythymidine-5'-Monophosphate
To be Published
|
|
4DWJ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate | | Descriptor: | THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate
To be Published
|
|
6BDZ
 
 | | ADAM10 Extracellular Domain Bound by the 11G2 Fab | | Descriptor: | 11G2 Fab Heavy Chain, 11G2 Fab Light Chain, CALCIUM ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BE6
 
 | | ADAM10 Extracellular Domain | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 10, SULFATE ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
