7U28
 
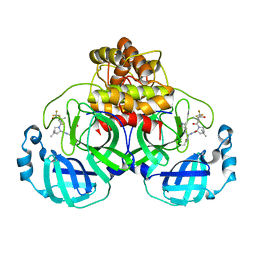 | | Structure of SARS-CoV-2 Mpro Lambda (G15S) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
4ZEG
 
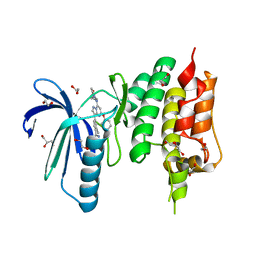 | | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2015-04-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of TTK kinase domain in complex with a pyrazolopyrimidine inhibitor.
To Be Published
|
|
3IED
 
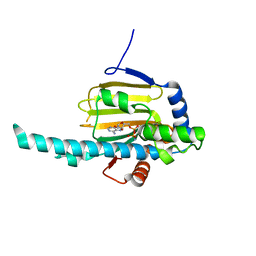 | | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN | | Descriptor: | AMP PHOSPHORAMIDATE, Heat shock protein | | Authors: | Pizarro, J.C, Wernimont, A.K, Lew, J, Hutchinson, A, Artz, J.D, Amaya, M.F, Plotnikova, O, Vedadi, M, Kozieradzki, I, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Botchkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of N-terminal domain of Plasmodium falciparum Hsp90 (PF14_0417) in complex with AMPPN
TO BE PUBLISHED
|
|
4O6L
 
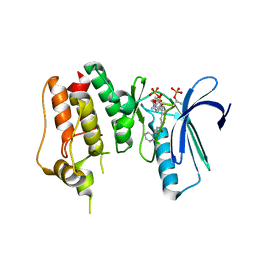 | | Crystal Structure of TTK kinase domain with an inhibitor: 401498 (N-[(1R)-1-(2-chlorophenyl)propyl]-3-{4-[(1-methylpiperidin-4-yl)oxy]phenyl}-1H-indazole-5-carboxamide) | | Descriptor: | Dual specificity protein kinase TTK, GLYCEROL, N-[(1R)-1-(2-chlorophenyl)propyl]-3-{4-[(1-methylpiperidin-4-yl)oxy]phenyl}-1H-indazole-5-carboxamide | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-12-22 | | Release date: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of TTK kinase domain with an inhibitor: 401498
TO BE PUBLISHED
|
|
1N3J
 
 | | Structure and Substrate of a Histone H3 Lysine Methyltransferase from Paramecium Bursaria Chlorella Virus 1 | | Descriptor: | Histone H3 Lysine Methyltransferase | | Authors: | Manzur, K.L, Farooq, A, Zeng, L, Plotnikova, O, Sachchidanand, Koch, A.W, Zhou, M.-M. | | Deposit date: | 2002-10-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric viral SET domain methyltransferase specific to Lys27 of histone H3.
Nat.Struct.Biol., 10, 2003
|
|
2RO1
 
 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
7TLL
 
 | | Structure of SARS-CoV-2 Mpro Omicron P132H in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Plotnikova, O, Liu, W, Stewart, A.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
1XR0
 
 | | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors | | Descriptor: | Basic fibroblast growth factor receptor 1, FGFR signalling adaptor SNT-1 | | Authors: | Dhalluin, C, Yan, K.S, Plotnikova, O, Lee, K.W, Zeng, L, Kuti, M, Mujtaba, S, Goldfarb, M.P, Zhou, M.-M. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of SNT PTB Domain Interactions with Distinct Neurotrophic Receptors
Mol.Cell, 6, 2000
|
|
1Y6Z
 
 | | Middle domain of Plasmodium falciparum putative heat shock protein PF14_0417 | | Descriptor: | heat shock protein, putative | | Authors: | Lunin, V.V, Botchkareva, E, Loppnau, P, Amani, M, Bray, J, Vedadi, M, Edwards, A, Arrowsmith, C, Sundstrom, M, Bochkarev, A, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-07 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4JS8
 
 | | Crystal structure of TTK kinase domain with an inhibitor: 401348 | | Descriptor: | 4-(cyclohexylmethoxy)-3-{4-[(1-methylpiperidin-4-yl)oxy]phenyl}-2H-indazole, DI(HYDROXYETHYL)ETHER, Dual specificity protein kinase TTK, ... | | Authors: | Qiu, W, Plotnikov, A.N, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of TTK kinase domain with an inhibitor: 401348
To be Published
|
|
4JXF
 
 | | Crystal Structure of PLK4 Kinase with an inhibitor: 400631 ((1R,2S)-2-{3-[(E)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}ETHENYL]-2H-INDAZOL-6-YL}-5'-METHOXYSPIRO[CYCLOPROPANE-1,3'-INDOL]-2'(1'H)-ONE) | | Descriptor: | (1R,2S)-2-{3-[(E)-2-{4-[(dimethylamino)methyl]phenyl}ethenyl]-2H-indazol-6-yl}-5'-methoxyspiro[cyclopropane-1,3'-indol]-2'(1'H)-one, 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK4 | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of PLK4 Kinase with an inhibitor: 400631
To be Published
|
|
2F6I
 
 | | Crystal structure of the ClpP protease catalytic domain from Plasmodium falciparum | | Descriptor: | ATP-dependent CLP protease, putative | | Authors: | Mulichak, A, Loppnau, P, Bray, J, Amani, M, Vedadi, M, Wasney, G, Finerty, P, Sundstrom, M, Weigelt, J, Edwards, A, Arrowsmith, C, Hui, R, Plotnikova, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum.
J.Mol.Biol., 404, 2010
|
|
7U29
 
 | | Structure of SARS-CoV-2 Mpro mutant (K90R) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2022-02-23 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis for the in vitro efficacy of nirmatrelvir against SARS-CoV-2 variants.
J.Biol.Chem., 298, 2022
|
|
3K8S
 
 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
1TXJ
 
 | | Crystal structure of translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi | | Descriptor: | translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi, PKN_PFE0545c | | Authors: | Walker, J.R, Vedadi, M, Sharma, S, Houston, S, Lew, J, Amani, M, Wasney, G, Skarina, T, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3PGG
 
 | | Crystal structure of cryptosporidium parvum u6 snrna-associated sm-like protein lsm5 | | Descriptor: | U6 snRNA-associated Sm-like protein LSm5. SM domain | | Authors: | Dong, A, Gao, M, Zhao, Y, Lew, J, Wasney, G.A, Kozieradzki, I, Vedadi, M, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-01 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Genome-Scale Protein Expression and Structural Biology of Plasmodium Falciparum and Related Apicomplexan Organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3TB2
 
 | | 1-Cys peroxidoxin from Plasmodium Yoelli | | Descriptor: | 1,2-ETHANEDIOL, 1-Cys peroxiredoxin, GLYCEROL, ... | | Authors: | Qiu, W, Artz, J.D, Vedadi, M, Sharma, S, Houston, S, Lew, J, Wasney, G, Amani, M, Xu, X, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-08-04 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1M3G
 
 | |
5WOF
 
 | | 1.65 ANGSTROM STRUCTURE OF THE DYNEIN LIGHT CHAIN 1 FROM PLASMODIUM FALCIPARUM | | Descriptor: | Dynein light chain 1, putative | | Authors: | Walker, J.R, Lew, J, Amani, M, Alam, Z, Wasney, G, Boulanger, K, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Hui, R, Botchkarev, A, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genome-scale Protein Expression and Structural Biology of
Plasmodium Falciparum and Related Apicomplexan Organisms.
MOL.BIOCHEM.PARASITOL., 151, 2007
|
|
5V8O
 
 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MPB
 
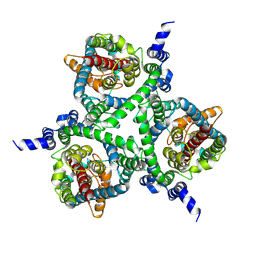 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6NAO
 
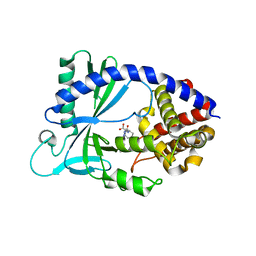 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | (1R,2S)-2-[(7-hydroxy-5-phenylpyrazolo[1,5-a]pyrimidine-3-carbonyl)amino]cyclohexane-1-carboxylic acid, CYCLIC GMP-AMP SYNTHASE, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2018-12-06 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
1HZM
 
 | |
1JSP
 
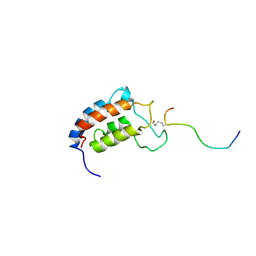 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
