2O72
 
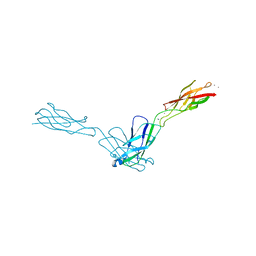 | | Crystal Structure Analysis of human E-cadherin (1-213) | | Descriptor: | CALCIUM ION, Epithelial-cadherin | | Authors: | Parisini, E, Wang, J.-H. | | Deposit date: | 2006-12-09 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human E-cadherin Domains 1 and 2, and Comparison with other Cadherins in the Context of Adhesion Mechanism
J.Mol.Biol., 373, 2007
|
|
1CKU
 
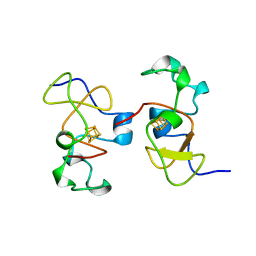 | | AB INITIO SOLUTION AND REFINEMENT OF TWO HIGH POTENTIAL IRON PROTEIN STRUCTURES AT ATOMIC RESOLUTION | | Descriptor: | IRON/SULFUR CLUSTER, PROTEIN (HIPIP) | | Authors: | Parisini, E, Capozzi, F, Lubini, P, Lamzin, V, Luchinat, C, Sheldrick, G.M. | | Deposit date: | 1999-04-24 | | Release date: | 1999-05-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ab initio solution and refinement of two high-potential iron protein structures at atomic resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6VGV
 
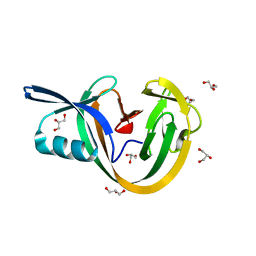 | | Crystal structure of VidaL intein | | Descriptor: | GLYCEROL, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VGW
 
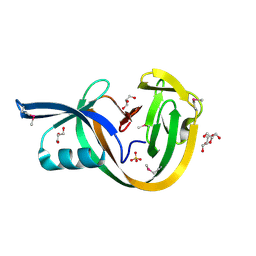 | | Crystal structure of VidaL intein (selenomethionine variant) | | Descriptor: | GLYCEROL, SULFATE ION, VidaL | | Authors: | Burton, A.J, Haugbro, M, Parisi, E, Muir, T.W. | | Deposit date: | 2020-01-09 | | Release date: | 2020-05-27 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Live-cell protein engineering with an ultra-short split intein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1M0G
 
 | | Solution structure of the alpha domain of mt_nc | | Descriptor: | CADMIUM ION, metallothionein MT_nc | | Authors: | Capasso, C, Carginale, V, Crescenzi, O, Di Maro, D, Parisi, E, Spadaccini, R, Temussi, P.A. | | Deposit date: | 2002-06-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MT_nc, a Novel Metallothionein from the Antarctic Fish Notothenia coriiceps.
Structure, 11, 2003
|
|
1M0J
 
 | | solution structure of the beta domain of mt_nc | | Descriptor: | CADMIUM ION, metallothionein MT_nc | | Authors: | Capasso, C, Carginale, V, Crescenzi, O, Di Maro, D, Parisi, E, Spadaccini, R, Temussi, P.A. | | Deposit date: | 2002-06-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of MT_nc, a Novel Metallothionein from the Antarctic Fish Notothenia coriiceps.
Structure, 11, 2003
|
|
4ZT1
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in x-dimer conformation | | Descriptor: | CALCIUM ION, Cadherin-1 | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4ZTE
 
 | | Crystal structure of human E-Cadherin (residues 3-213) in complex with a peptidomimetic inhibitor | | Descriptor: | CALCIUM ION, Cadherin-1, N-{[(2S,5S)-1-benzyl-5-(2-{[(2S,3S)-1-(tert-butylamino)-3-methyl-1-oxopentan-2-yl]amino}-2-oxoethyl)-3,6-dioxopiperazin-2-yl]methyl}-L-alpha-asparagine | | Authors: | Nardone, V, Lucarelli, A.P, Dalle Vedove, A, Parisini, E. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of Human E-Cadherin-EC1EC2 in Complex with a Peptidomimetic Competitive Inhibitor of Cadherin Homophilic Interaction.
J.Med.Chem., 59, 2016
|
|
4WCT
 
 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2014-09-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
8OU8
 
 | | Crystal structure of E. coli threonyl tRNA synthetase in complex with a TM84 analogue | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Threonine--tRNA ligase, ... | | Authors: | Rodriguez Buitrago, J.A, Parisini, E, Aigars, J. | | Deposit date: | 2023-04-22 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis and evaluation of an agrocin 84 toxic moiety (TM84) analogue as a malarial threonyl tRNA synthetase inhibitor.
Org.Biomol.Chem., 21, 2023
|
|
6TOR
 
 | | human O-phosphoethanolamine phospho-lyase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Ethanolamine-phosphate phospho-lyase, GLYCEROL | | Authors: | Vettraino, C, Donini, S, Parisini, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of human O-phosphoethanolamine phospho-lyase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
1BLX
 
 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
8QIC
 
 | |
6QS9
 
 | |
8CMV
 
 | |
4XWZ
 
 | | The crystal structure of Fructosyl amine: oxygen oxidoreductase (Amadoriase I) from Aspergillus fumigatus in complex with the substrate fructosyl lysine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl amine:oxygen oxidoreductase, LYSINE, ... | | Authors: | Rigoldi, F, Gautieri, A, Dalle Vedove, A, Lucarelli, A.P, Vesentini, S, Parisini, E. | | Deposit date: | 2015-01-29 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the deglycating enzyme Amadoriase I in its free form and substrate-bound complex.
Proteins, 84, 2016
|
|
7B9H
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-42a | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{E})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-12-14 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
7AY6
 
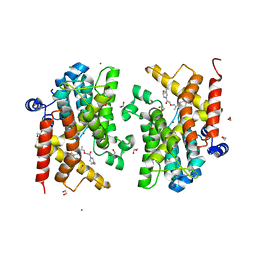 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-41b | | Descriptor: | 1,2-ETHANEDIOL, 2-[(~{Z})-1-(3-cyclopentyloxy-4-methoxy-phenyl)ethylideneamino]oxy-1-[(2~{S},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ... | | Authors: | Torretta, A, Abbate, S, Parisini, E. | | Deposit date: | 2020-11-11 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Design, synthesis, biological evaluation and structural characterization of novel GEBR library PDE4D inhibitors.
Eur.J.Med.Chem., 223, 2021
|
|
2ATP
 
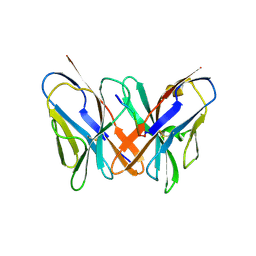 | | Crystal structure of a CD8ab heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD8 alpha chain, T-cell surface glycoprotein CD8 beta chain, ... | | Authors: | Chang, H.C, Tan, K, Ouyang, J, Parisini, E, Liu, J.H, Le, Y, Wang, X, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2005-08-25 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of a CD8alphabeta Heterodimer and Comparison with the CD8alphaalpha Homodimer.
Immunity, 23, 2005
|
|
6Y4J
 
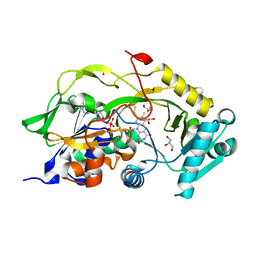 | | Engineered Fructosyl Peptide Oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fructosyl Peptide Oxidase, GLYCEROL, ... | | Authors: | Donini, S, Rigoldi, F, Gautieri, A, Parisini, E. | | Deposit date: | 2020-02-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Rational backbone redesign of a fructosyl peptide oxidase to widen its active site access tunnel.
Biotechnol.Bioeng., 117, 2020
|
|
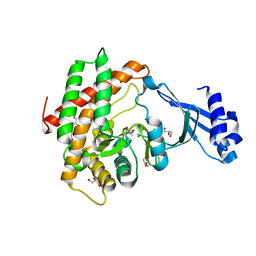
6YXS
 
 | |
6YXT
 
 | |
4OY9
 
 | | Crystal structure of human P-Cadherin EC1-EC2 in closed conformation | | Descriptor: | CALCIUM ION, Cadherin-3 | | Authors: | Dalle Vedove, A, Lucarelli, A.P, Nardone, V, Matino, A, Parisini, E. | | Deposit date: | 2014-02-11 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The X-ray structure of human P-cadherin EC1-EC2 in a closed conformation provides insight into the type I cadherin dimerization pathway.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6F8W
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-18a | | Descriptor: | 3-[3-(3-cyclopentyloxy-4-methoxy-phenyl)pyrazol-1-yl]-1-morpholin-4-yl-propan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
6F6U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-7b | | Descriptor: | 2-[(~{E})-(3-cyclopentyloxy-4-methoxy-phenyl)methylideneamino]oxy-1-[(2~{R},6~{S})-2,6-dimethylmorpholin-4-yl]ethanone, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
