6MVM
 
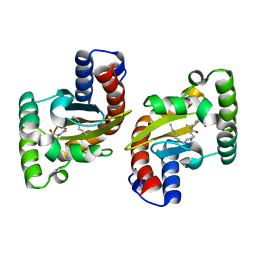 | | LasR LBD L130F:3OC14HSL complex | | Descriptor: | 3-oxo-N-[(3S)-2-oxooxolan-3-yl]tetradecanamide, Transcriptional regulator LasR | | Authors: | Paczkowski, J.E, Bassler, B.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
4Q66
 
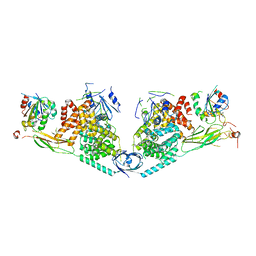 | | Structure of Exomer bound to Arf1. | | Descriptor: | ADP-ribosylation factor 1, Chs5p, MAGNESIUM ION, ... | | Authors: | Paczkowski, J.E, Fromme, J.C. | | Deposit date: | 2014-04-21 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | Structural basis for membrane binding and remodeling by the exomer secretory vesicle cargo adaptor.
Dev.Cell, 30, 2014
|
|
4YG8
 
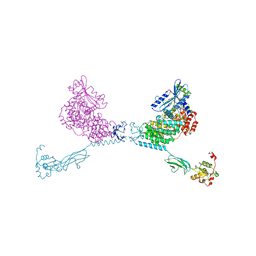 | | CRYSTAL STRUCTURE OF THE CHS5-CHS6 EXOMER CARGO ADAPTOR COMPLEX | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Chitin biosynthesis protein CHS5, Chitin biosynthesis protein CHS6, ... | | Authors: | Paczkowski, J.E, Richardson, B.C, Strassner, A.M, Fromme, J.C. | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The exomer cargo adaptor structure reveals a novel GTPase-binding domain.
Embo J., 31, 2012
|
|
6UGL
 
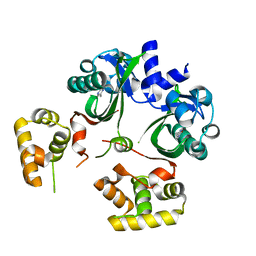 | | VqmA bound to DPO | | Descriptor: | 3,5-dimethylpyrazin-2(1H)-one, Helix-turn-helix transcriptional regulator | | Authors: | Paczkowski, J.E, Huang, X. | | Deposit date: | 2019-09-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism underlying autoinducer recognition in theVibrio choleraeDPO-VqmA quorum-sensing pathway.
J.Biol.Chem., 295, 2020
|
|
8DQ0
 
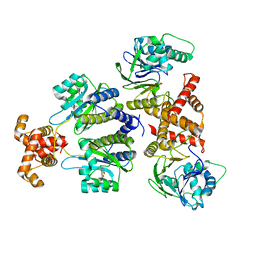 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, RhlR protein | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
8DQ1
 
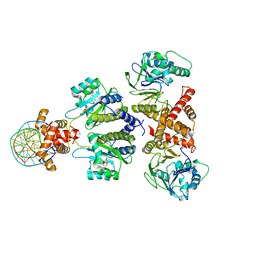 | | Quorum-sensing receptor RhlR bound to PqsE | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, DNA (5'-D(*AP*CP*CP*TP*GP*CP*CP*AP*GP*AP*CP*TP*GP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Paczkowski, J.E, Fromme, J.C, Feathers, J.R. | | Deposit date: | 2022-07-18 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the RhlR-PqsE complex from Pseudomonas aeruginosa reveals mechanistic insights into quorum-sensing gene regulation.
Structure, 30, 2022
|
|
6MWW
 
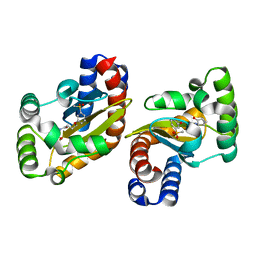 | | LasR LBD:BB0126 complex | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWL
 
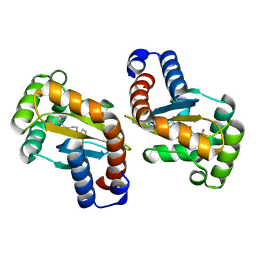 | | LasR LBD:mBTL complex | | Descriptor: | 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWH
 
 | | LasR LBD:BB0020 complex | | Descriptor: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MVN
 
 | | LasR LBD L130F:3OC10HSL complex | | Descriptor: | 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants driving homoserine lactone ligand selection in thePseudomonas aeruginosaLasR quorum-sensing receptor.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7KGX
 
 | | Structure of PQS Response Protein PqsE in Complex with 4-(3-(2-methyl-2-morpholinobutyl)ureido)-N-(thiazol-2-yl)benzamide | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-({[(2R)-2-methyl-2-(morpholin-4-yl)butyl]carbamoyl}amino)-N-(1,3-thiazol-2-yl)benzamide, FE (III) ION | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production.
Acs Chem.Biol., 16, 2021
|
|
7KGW
 
 | |
