6PRF
 
 | | HIV-1 Protease multiple drug resistant clinical isolate mutant PR20 with GRL-14213A | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease, YTTRIUM ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6P9B
 
 | |
6P9A
 
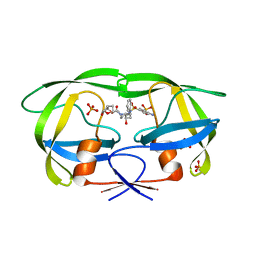 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
6WQF
 
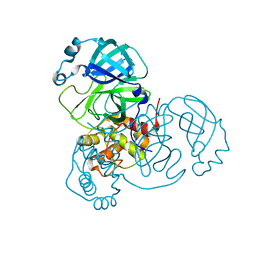 | |
6XQS
 
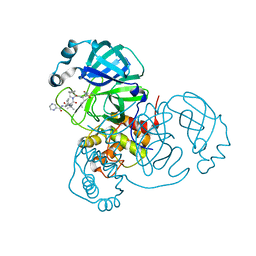 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
6XB2
 
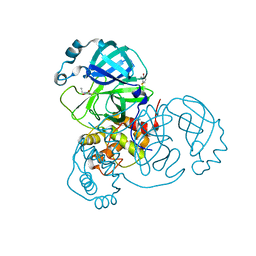 | | Room temperature X-ray crystallography reveals catalytic cysteine in the SARS-CoV-2 3CL Mpro is highly reactive: Insights for enzyme mechanism and drug design | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL M pro : insights into enzyme mechanism and drug design.
Iucrj, 7, 2020
|
|
6XB0
 
 | |
6XQU
 
 | |
6XHU
 
 | |
6XB1
 
 | |
6XCH
 
 | |
6XQT
 
 | | Room-temperature X-ray Crystal structure of SARS-CoV-2 main protease in complex with Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kneller, D.W, Kovalevsky, A, Coates, L. | | Deposit date: | 2020-07-10 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Structure, 28, 2020
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | Descriptor: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | Descriptor: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | Descriptor: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | Descriptor: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RN4
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMZ
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-63 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7N8C
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule5948770040 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperazin-1-yl]carbonyl-1~{H}-pyrimidine-2,4-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-06-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7N89
 
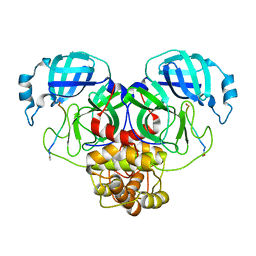 | |
7SI9
 
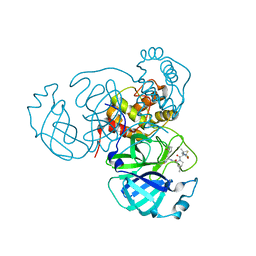 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
