2AXW
 
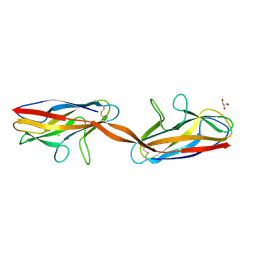 | | Structure of DraD invasin from uropathogenic Escherichia coli | | Descriptor: | CHLORIDE ION, DraD invasin, GLYCEROL | | Authors: | Jedrzejczak, R, Dauter, Z, Dauter, M, Piatek, R, Zalewska, B, Mroz, M, Bury, K, Nowicki, B, Kur, J. | | Deposit date: | 2005-09-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped beta-tails.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2FXQ
 
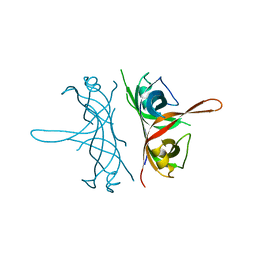 | |
3RC3
 
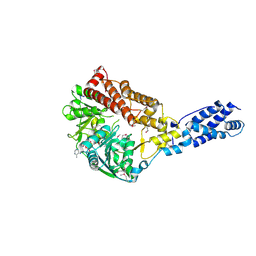 | | Human Mitochondrial Helicase Suv3 | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, AZIDE ION, ... | | Authors: | Dauter, Z, Jedrzejczak, R, Dauter, M, Szczesny, R, Stepien, P. | | Deposit date: | 2011-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3RC8
 
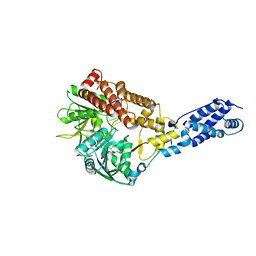 | | Human Mitochondrial Helicase Suv3 in Complex with Short RNA Fragment | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, RNA fragment | | Authors: | Dauter, Z, Jedrzejczak, R, Dauter, M, Wang, J, Szczesny, R, Stepien, P. | | Deposit date: | 2011-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3K1U
 
 | | Beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-xylosidase, ... | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-06 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of
beta-xylosidase, family 43 glycosyl hydrolase from Clostridium acetobutylicum at 1.55 A resolution
To be Published
|
|
5JBR
 
 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | Descriptor: | SULFATE ION, Uncharacterized protein Bcav_2135 | | Authors: | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-04-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
7M5F
 
 | | Contact-dependent inhibition system from Serratia marcescens BWH57 | | Descriptor: | CdiI, MALONATE ION, Toxin CdiA | | Authors: | Michalska, K, Nutt, W, Stols, L, Jedrzejczak, R, Hayes, C.S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Contact-dependent inhibition system from Serratia marcescens
To Be Published
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
4N04
 
 | |
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
4MQB
 
 | | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TETRAETHYLENE GLYCOL, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of thymidylate kinase from Staphylococcus aureus in complex with 2-(N-morpholino)ethanesulfonic acid
To be Published
|
|
4MOZ
 
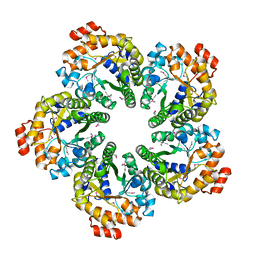 | |
4WHI
 
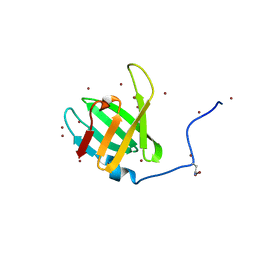 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | Descriptor: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
8GHX
 
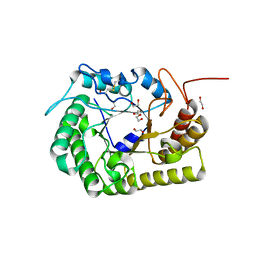 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHY
 
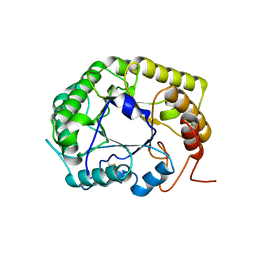 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
5T86
 
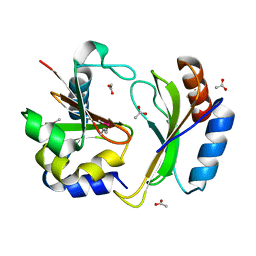 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
7RL8
 
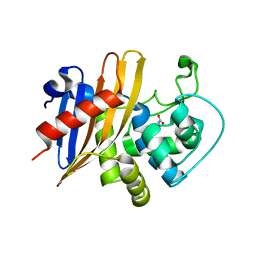 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RLR
 
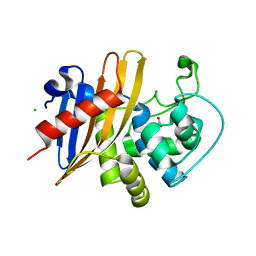 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RBS
 
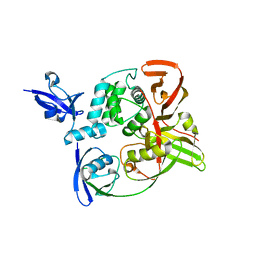 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with human ISG15 | | Descriptor: | Papain-like protease, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7N3C
 
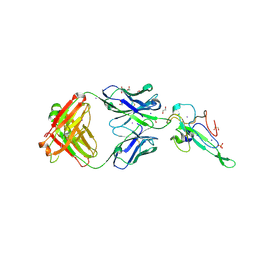 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7N3D
 
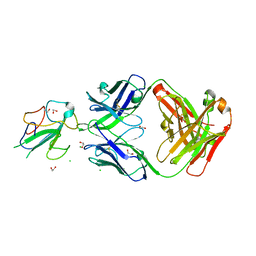 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
6UAG
 
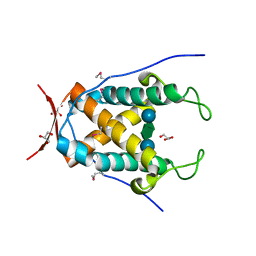 | |
6UG4
 
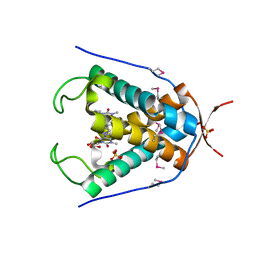 | |
6UHS
 
 | |
