8PMI
 
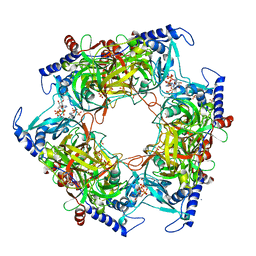 | | Structure of Nal1 indica cultivar IR64, construct 36-458 in presence of peptide from FZP protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AMMONIUM ION, MAGNESIUM ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PME
 
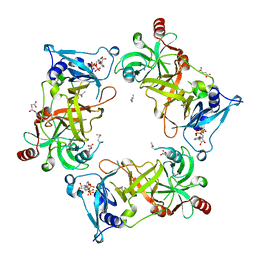 | | Structure of Nal1 indica cultivar IR64, construct 88-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Narrow leaf 1, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PML
 
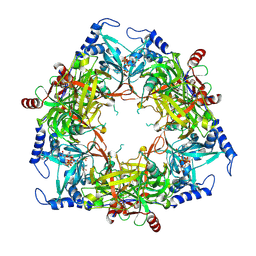 | | Structure of Nal1 protein , SPIKE allele from japonica rice, construct 46-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMG
 
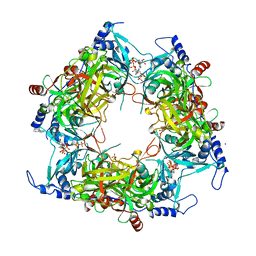 | | Structure of Nal1 indica cultivar IR64, construct 36-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMM
 
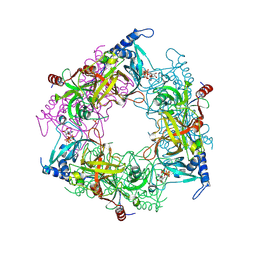 | | Structure of Nal1 protein, allele SPIKE from japonica rice, construct 31-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PN2
 
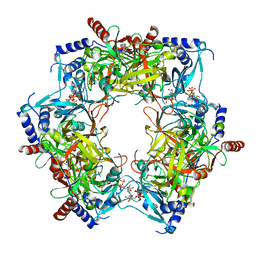 | | CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PN1
 
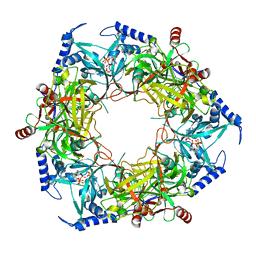 | | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
5EGC
 
 | |
6N48
 
 | | Structure of beta2 adrenergic receptor bound to BI167107, Nanobody 6B9, and a positive allosteric modulator | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid Antibody Fragment, ... | | Authors: | Liu, X, Masoudi, A, Kahsai, A.W, Huang, L.Y, Pani, B, Hirata, K, Ahn, S, Lefkowitz, R.J, Kobilka, B.K. | | Deposit date: | 2018-11-17 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of beta2AR regulation by an intracellular positive allosteric modulator.
Science, 364, 2019
|
|
5X8I
 
 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
6J3P
 
 | | Crystal structure of the human GCN5 bromodomain in complex with compound (R,R)-36n | | Descriptor: | 2-{[(3R,5R)-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-methylpiperidin-3-yl]amino}-3-methyl-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2A | | Authors: | Huang, L.Y, Li, H, Niu, L, Wu, C.Y, Yu, Y.M, Li, L.L, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
4JQI
 
 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
6NI2
 
 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NI3
 
 | | B2V2R-Gs protein subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Endolysin,Beta-2 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5LY0
 
 | | Crystal structure of LOB domain of Ramosa2 from Wheat | | Descriptor: | LOB family transfactor Ramosa2.1, ZINC ION | | Authors: | Wei, X.-B, Zhang, B, Chen, W.-F, Fan, S.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-23 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural analysis reveals a "molecular calipers" mechanism for a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor protein from wheat.
J. Biol. Chem., 294, 2019
|
|
8PO7
 
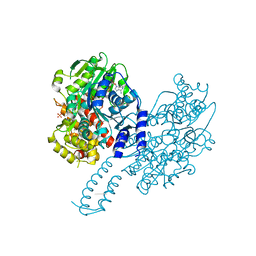 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO8
 
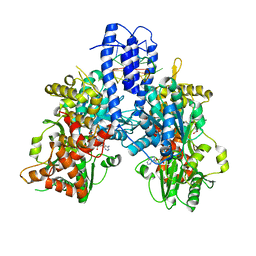 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO6
 
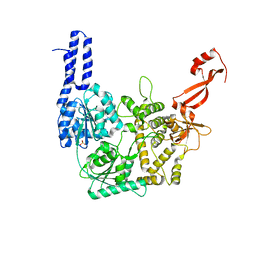 | | Structure of Escherichia coli HrpA apo form | | Descriptor: | ATP-dependent RNA helicase HrpA, PHOSPHATE ION | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
