1YKQ
 
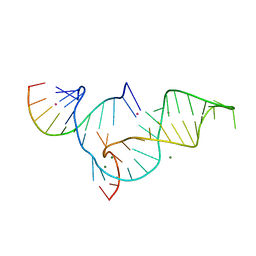 | | Crystal structure of Diels-Alder ribozyme | | Descriptor: | CADMIUM ION, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YKV
 
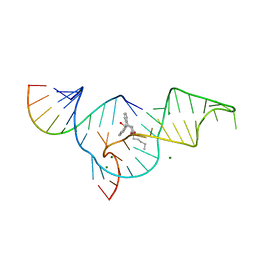 | | Crystal structure of the Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, Diels-Alder ribozyme, MAGNESIUM ION | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-18 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1YLS
 
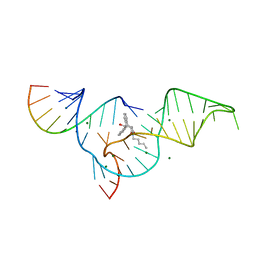 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7OZV
 
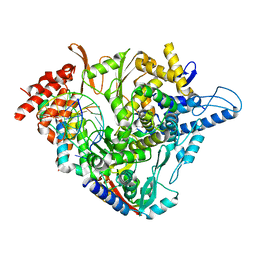 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7OZU
 
 | | SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with A | | Descriptor: | Non-structural protein 7, Non-structural protein 8, Product RNA, ... | | Authors: | Kabinger, F, Stiller, C, Schmitzova, J, Dienemann, C, Kokic, G, Hillen, H.S, Hoebartner, C, Cramer, P. | | Deposit date: | 2021-06-28 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2H1M
 
 | |
8REV
 
 | | Structure of XPD stalled at a Y-fork DNA containing a interstrand crosslink | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase CHL1, DNA (46-MER), ... | | Authors: | Kuper, J, Hove, T, Kisker, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | XPD stalled on cross-linked DNA provides insight into damage verification.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7OA3
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium hexammine co-crystallized form) | | Descriptor: | Chili RNA Aptamer, DMHBO+, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-19 | | Release date: | 2021-06-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAX
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBO+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAW
 
 | | Crystal structure of the Chili RNA aptamer in complex with DMHBI+ | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBI+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7OAV
 
 | | Crystal structure of Chili RNA aptamer in complex with DMHBO+ (Iridium III hexammine soaking crystal form) | | Descriptor: | CHLORIDE ION, Chili RNA Aptamer, DMHBO+, ... | | Authors: | Mieczkowski, M, Pena, V, Hoebartner, C. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Large Stokes shift fluorescence activation in an RNA aptamer by intermolecular proton transfer to guanine.
Nat Commun, 12, 2021
|
|
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3B
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -3 (structure 1) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3C
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
5DUN
 
 | | The crystal structure of OMe substituted twister ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | Authors: | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | Deposit date: | 2015-09-19 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5CKI
 
 | | Crystal structure of 9DB1* deoxyribozyme (Cobalt hexammine soaked crystals) | | Descriptor: | COBALT (II) ION, DNA (44-MER), MAGNESIUM ION, ... | | Authors: | Ponce-Salvatierra, A, Hoebartner, C, Pena, V. | | Deposit date: | 2015-07-15 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.985 Å) | | Cite: | Crystal structure of a DNA catalyst.
Nature, 529, 2016
|
|
5CKK
 
 | |
7Q7Z
 
 | |
7Q81
 
 | |
7Q7X
 
 | |
7Q80
 
 | |
7Q82
 
 | |
7Q7Y
 
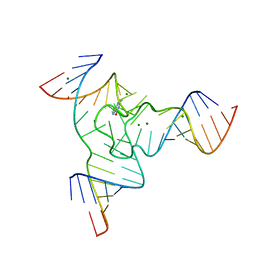 | |
1Y27
 
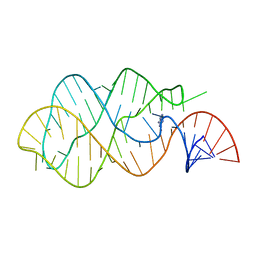 | | G-riboswitch-guanine complex | | Descriptor: | Bacillus subtilis xpt, GUANINE | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
1Y26
 
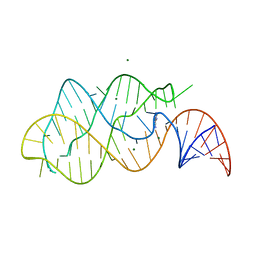 | | A-riboswitch-adenine complex | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus A-riboswitch | | Authors: | Serganov, A, Yuan, Y.R, Patel, D.J. | | Deposit date: | 2004-11-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs
Chem.Biol., 11, 2004
|
|
