1IK4
 
 | | X-ray Structure of Methylglyoxal Synthase from E. coli Complexed with Phosphoglycolohydroxamic Acid | | Descriptor: | METHYLGLYOXAL SYNTHASE, PHOSPHOGLYCOLOHYDROXAMIC ACID | | Authors: | Marks, G.T, Harris, T.K, Massiah, M.A, Mildvan, A.S, Harrison, D.H.T. | | Deposit date: | 2001-05-02 | | Release date: | 2001-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic implications of methylglyoxal synthase complexed with phosphoglycolohydroxamic acid as observed by X-ray crystallography and NMR spectroscopy.
Biochemistry, 40, 2001
|
|
7NBG
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z52314092, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBH
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z26781964, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBD
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z235449082, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBC
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z2856434779, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
7NBF
 
 | | Crystal structure of human serine racemase in complex with DSiP fragment Z126932614, XChem fragment screen. | | Descriptor: | 1,2-ETHANEDIOL, 2-[(methylsulfonyl)methyl]-1H-benzimidazole, CALCIUM ION, ... | | Authors: | Koulouris, C.R, Roe, S.M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZSP
 
 | | Human serine racemase bound to ATP and malonate. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-16 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6ZUJ
 
 | | Human serine racemase holoenzyme from 20% DMSO soak (XChem crystallographic fragment screen). | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koulouris, C.R, Bax, B.D, Roe, S.M, Atack, J.R. | | Deposit date: | 2020-07-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Commun Biol, 5, 2022
|
|
6SM0
 
 | | Venus 66 p-Azido-L-Phenylalanin (azF) variant, dark grown | | Descriptor: | Green fluorescent protein, OXYGEN MOLECULE, SULFATE ION, ... | | Authors: | Rizkallah, P.J, Al Maslookhi, H.S, Jones, D.D. | | Deposit date: | 2019-08-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Stalling chromophore synthesis of the fluorescent protein Venus reveals the molecular basis of the final oxidation step.
Chem Sci, 12, 2021
|
|
4E4L
 
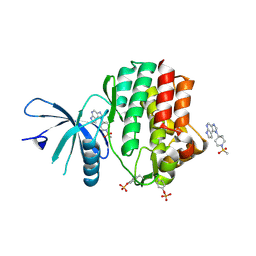 | | JAK1 kinase (JH1 domain) in complex with compound 30 | | Descriptor: | 1-[4-methyl-1-(methylsulfonyl)piperidin-4-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-30 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E4M
 
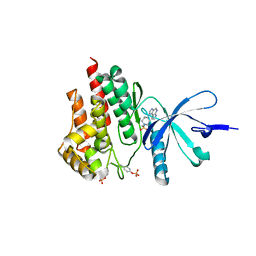 | | JAK2 kinase (JH1 domain) in complex with compound 30 | | Descriptor: | 1-[4-methyl-1-(methylsulfonyl)piperidin-4-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E5W
 
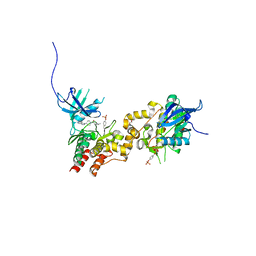 | | JAK1 kinase (JH1 domain) in complex with compound 26 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine-protein kinase JAK1, [4-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)piperidin-1-yl][(2S)-1-(propan-2-yl)pyrrolidin-2-yl]methanone | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-14 | | Release date: | 2012-05-30 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E6D
 
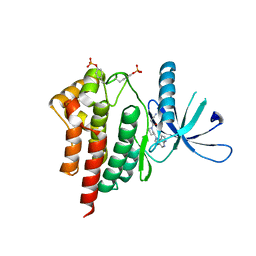 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 7 | | Descriptor: | 3-[(3R)-3-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)piperidin-1-yl]-3-oxopropanenitrile, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2023-03-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E4N
 
 | | JAK1 kinase (JH1 domain) in complex with compound 49 | | Descriptor: | Tyrosine-protein kinase JAK1, tert-butyl [(1R,3R)-3-(imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl)cyclopentyl]carbamate | | Authors: | Eigenbrot, C. | | Deposit date: | 2012-03-13 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
4E6Q
 
 | | JAK2 kinase (JH1 domain) triple mutant in complex with compound 12 | | Descriptor: | 1-(1-benzylpiperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK2 | | Authors: | Murray, J.M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-05-30 | | Last modified: | 2012-07-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Identification of Imidazo-Pyrrolopyridines as Novel and Potent JAK1 Inhibitors.
J.Med.Chem., 55, 2012
|
|
