7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
7BQ5
 
 | | ZIKV sE bound to mAb Z6 | | Descriptor: | Z6 Light Chain, Z6 heavy chain, envelope protein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
7BPK
 
 | | Zika virus envelope protein mutant bound to mAb | | Descriptor: | Envelope protein, IG c307_light_IGLV1-51_IGLJ2, Z3L1 Heavy chain | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protective Zika vaccines engineered to eliminate enhancement of dengue infection via immunodominance switch.
Nat.Immunol., 22, 2021
|
|
3JTT
 
 | |
3JTS
 
 | | GY9-Mamu-A*02-hb2m | | Descriptor: | Beta-2-microglobulin, MHC class I Mamu-A*02, peptide of Gag-Pol polyprotein | | Authors: | Dai, L, Feng, Y, Qi, J, Gao, G.F. | | Deposit date: | 2009-09-14 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of Mamu A*2
To be Published
|
|
5JHL
 
 | | Crystal structure of zika virus envelope protein in complex with a flavivirus broadly-protective antibody | | Descriptor: | Antibody Heavy chain, antibody Light chain, envelope protein | | Authors: | Dai, L, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody.
Cell Host Microbe, 19, 2016
|
|
5JHM
 
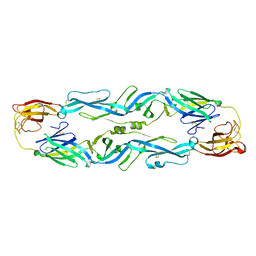 | |
6A7U
 
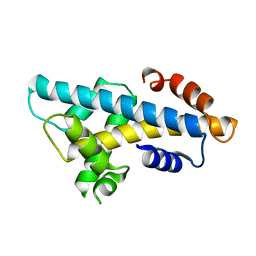 | | Crystal structure of histone H2A.Bbd-H2B dimer | | Descriptor: | Histone H2B type 2-E,Histone H2A-Bbd type 2/3 | | Authors: | Dai, L, Zhou, Z. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the histone heterodimer containing histone variant H2A.Bbd.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7FIR
 
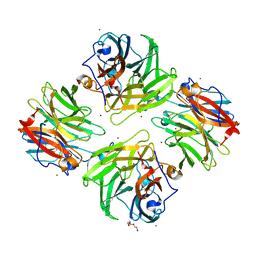 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
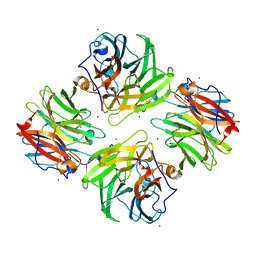 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
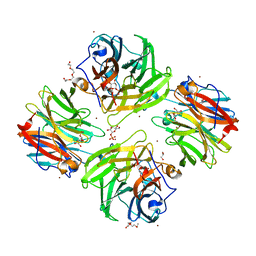 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7XWM
 
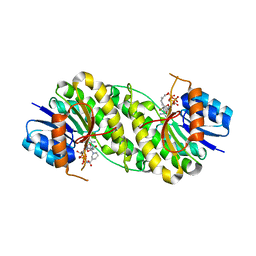 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWJ
 
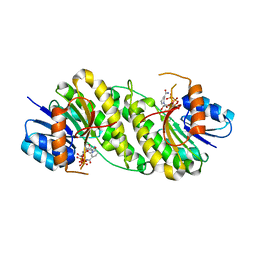 | | structure of patulin-detoxifying enzyme Y155F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWI
 
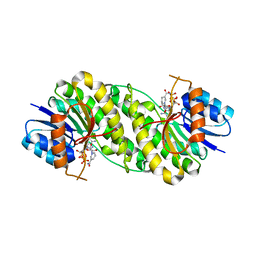 | | structure of patulin-detoxifying enzyme with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWH
 
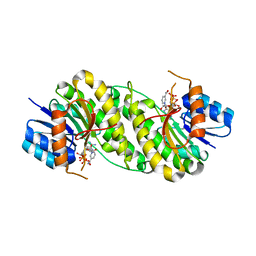 | | structure of patulin-detoxifying enzyme with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWL
 
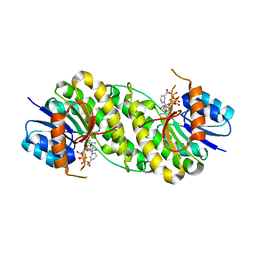 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWN
 
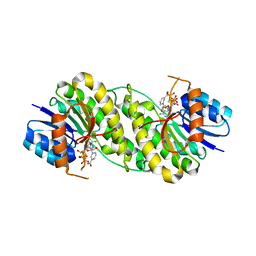 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
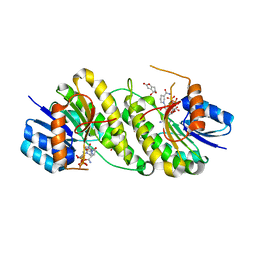 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
8HF8
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist V1 | | Descriptor: | 2-[4-[[2,5-bis(oxidanylidene)-3-[4-(trifluoromethyl)phenyl]imidazolidin-1-yl]methyl]-2,6-dimethyl-phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of the First Subnanomolar PPAR alpha / delta Dual Agonist for the Treatment of Cholestatic Liver Diseases.
J.Med.Chem., 66, 2023
|
|
7W0G
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | Descriptor: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
7E8I
 
 | | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (145-MER), Histone H2A, ... | | Authors: | Dai, Y, Dai, L, Zhou, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin.
Mol.Cell, 81, 2021
|
|
8JZ4
 
 | | Crystal structure of AetF in complex with FAD and 5-bromo-L-tryptophan | | Descriptor: | 5-bromo-L-tryptophan, AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ2
 
 | | Crystal structure of AetF in complex with FAD | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
8JZ3
 
 | | Crystal structure of AetF in complex with FAD and L-tryptophan | | Descriptor: | AetF, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN | | Authors: | Li, H, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into the self-sufficient flavin-dependent halogenase.
Int.J.Biol.Macromol., 260, 2024
|
|
