8F2T
 
 | |
8F2V
 
 | |
2GVX
 
 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
9EWK
 
 | | Solvent organization in ultrahigh-resolution protein crystal structure at room temperature | | Descriptor: | Crambin, ETHANOL | | Authors: | Chen, J.C.-H, Gilski, M, Chang, C, Borek, D, Rosenbaum, G, Lavens, A, Otwinowski, Z, Kubicki, M, Dauter, Z, Jaskolski, M, Joachimiak, A. | | Deposit date: | 2024-04-04 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.7 Å) | | Cite: | Solvent organization in the ultrahigh-resolution crystal structure of crambin at room temperature.
Iucrj, 11, 2024
|
|
1EXQ
 
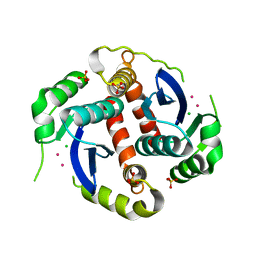 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EX4
 
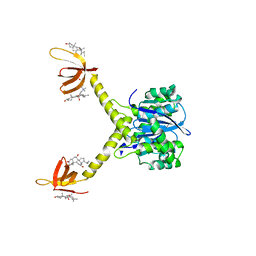 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2BCE
 
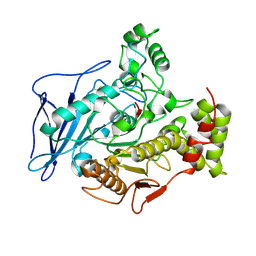 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
3U7T
 
 | |
3HLI
 
 | |
3HLH
 
 | |
3LI5
 
 | | Diisopropyl fluorophosphatase (DFPase), E21Q,N120D,N175D,D229N mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3LI4
 
 | | Diisopropyl fluorophosphatase (DFPase), N120D,N175D,D229N mutant | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3LI3
 
 | | Diisopropyl fluorophosphatase (DFPase), D121E mutant | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5J04
 
 | |
1OSF
 
 | | Human Hsp90 in complex with 17-desmethoxy-17-N,N-Dimethylaminoethylamino-Geldanamycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 17-DESMETHOXY-17-N,N-DIMETHYLAMINOETHYLAMINO-GELDANAMYCIN, ACETIC ACID, ... | | Authors: | Jez, J.M, Chen, J.C.-H, Rastelli, G, Stroud, R.M, Santi, D.V. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Molecular Modeling of 17-DMAG in Complex with Human Hsp90
Chem.Biol., 10, 2003
|
|
2NUB
 
 | | Structure of Aquifex aeolicus Argonuate | | Descriptor: | Argonaute | | Authors: | Rashid, U.J, Paterok, D, Koglin, A, Gohlke, H, Piehler, J, Chen, J.C.-H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function.
J.Biol.Chem., 282, 2007
|
|
4FC1
 
 | |
3BYC
 
 | | Joint neutron and X-ray structure of diisopropyl fluorophosphatase. Deuterium occupancies are 1-Q, where Q is occupancy of H | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Mustyakimov, M, Ruterjans, H, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Rapid determination of hydrogen positions and protonation states of diisopropyl fluorophosphatase by joint neutron and X-ray diffraction refinement.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3KGG
 
 | | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): Perdeuteration of proteins for neutron diffraction | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Tomanicek, S.J, John, H, Hanson, B.L, terjans, H.R, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): perdeuteration of proteins for neutron diffraction.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
