8UEL
 
 | | Crystal structure of enolase from Litopenaeus vannamei | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Chang, X, Zhao, G. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and Structural Analyses of Enolase from Shrimp ( Litopenaeus vannamei ).
J.Agric.Food Chem., 71, 2023
|
|
1AI0
 
 | | R6 HUMAN INSULIN HEXAMER (NON-SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
1AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
1D0R
 
 | |
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'GREEN' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
5AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'RED' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
2AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 20 STRUCTURES | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-28 | | Release date: | 2000-02-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, REFINED AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
9L3Z
 
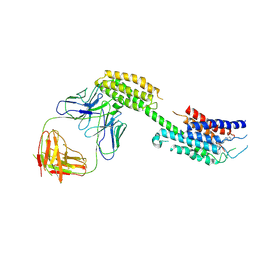 | | Cryo-EM structure of the inactive chemokine-like receptor 1 (CMKLR1) | | Descriptor: | Chemerin-like receptor 1,Soluble cytochrome b562, LRH7-C2, The heavy chain of anti-BRIL Fab, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3W
 
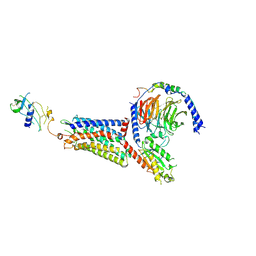 | | Cryo-EM structure of the chemokine-like receptor 1 in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
9L3Y
 
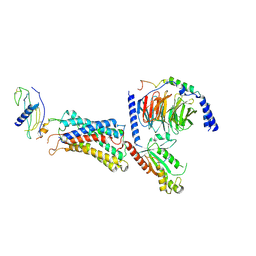 | | Cryo-EM structure of the G-protein coupled receptor 1 (GPR1) in complex with chemerin and Gi1 | | Descriptor: | Chemerin-like receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, He, M, Wu, B, Zhao, Q. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the distinct ligand recognition and signaling of the chemerin receptors CMKLR1 and GPR1.
Protein Cell, 16, 2025
|
|
7ES4
 
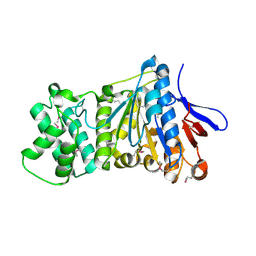 | | the crystral structure of DndH-C-domain | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptH | | Authors: | Wu, D. | | Deposit date: | 2021-05-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
7EXX
 
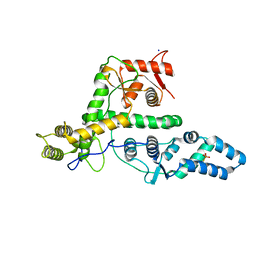 | | The structure of DndG | | Descriptor: | DNA phosphorothioation-dependent restriction protein DptG, SODIUM ION | | Authors: | Wu, D, Wang, L. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The functional coupling between restriction and DNA phosphorothioate modification systems underlying the DndFGH restriction complex
Nat Catal, 2022
|
|
9L09
 
 | | SARS-CoV-2 C-RTC with 13-TP | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Huang, Y.C, Liang, L, Liu, Y.X, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2024-12-12 | | Release date: | 2025-03-19 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a 2'-a-Fluoro-2'-b-C-(fluoromethyl) Purine Nucleotide Prodrug as a Potential Oral Anti-SARS-CoV-2 Agent
J Med Chem, 68, 2025
|
|
8WET
 
 | | The cryo-EM structure of TdpAB complex | | Descriptor: | TdpA, TdpB | | Authors: | An, T, Tan, Q. | | Deposit date: | 2023-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
Nat.Chem.Biol., 2025
|
|
8WFD
 
 | | The cryo-EM structure of TdpAB in complex with AMPPNP and DNA | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*TP*TP*TP*TP*GP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*CP*AP*AP*AP*AP*GP*GP*GP*C)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | An, T, Tan, Q. | | Deposit date: | 2023-09-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
Nat.Chem.Biol., 2025
|
|
8ZF0
 
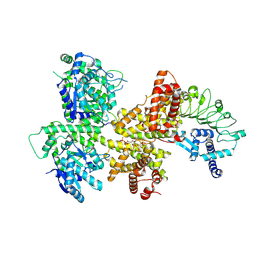 | | pRib-ADP bound OsEDS1-PAD4-ADR1 complex | | Descriptor: | EDS1, Lipase-like PAD4, RPW8 domain-containing protein, ... | | Authors: | Xu, W.Y, Zhang, Y. | | Deposit date: | 2024-05-07 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A canonical protein complex controls immune homeostasis and multipathogen resistance.
Science, 386, 2024
|
|
8Y1K
 
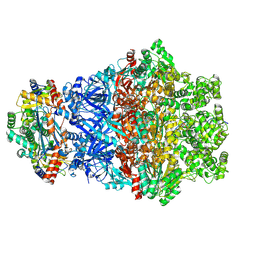 | | The cryo-EM structure of TdpAB in complex with AMPPNP and PT-DNA | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*TP*TP*TP*TP*GP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*CP*AP*AP*AP*AP*GP*GP*GP*C)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, T.A, Qian, T. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
Nat.Chem.Biol., 2025
|
|
7X9X
 
 | | Thermotoga Maritima ferritin variant-Tm-E(G40E) with Zn | | Descriptor: | FE (III) ION, Ferritin, ZINC ION | | Authors: | Liu, Y, Zhao, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Directed Self-Assembly of Dimeric Building Blocks into Networklike Protein Origami to Construct Hydrogels.
Acs Nano, 16, 2022
|
|
7XA4
 
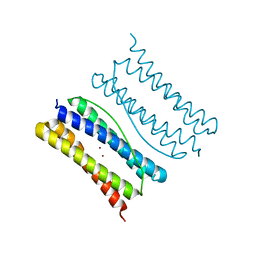 | |
7XA2
 
 | | Thermotoga maritima ferritin variant-Tm-E(S111H) | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Liu, Y, Zhao, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Directed Self-Assembly of Dimeric Building Blocks into Networklike Protein Origami to Construct Hydrogels.
Acs Nano, 16, 2022
|
|
8KD5
 
 | | Rpd3S in complex with nucleosome with H3K36MLA modification and 187bp DNA, class2 | | Descriptor: | 187bp DNA, Chromatin modification-related protein EAF3, Histone H2A, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Chang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
