1RKT
 
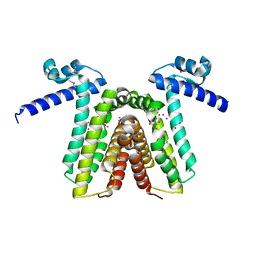 | | Crystal structure of yfiR, a putative transcriptional regulator from Bacillus subtilis | | Descriptor: | UNKNOWN ATOM OR ION, protein yfiR | | Authors: | Anderson, W.F, Rajan, S.S, Yang, X, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-23 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of YfiR, an unusual TetR/CamR-type putative transcriptional regulator from Bacillus subtilis.
Proteins, 65, 2006
|
|
4LES
 
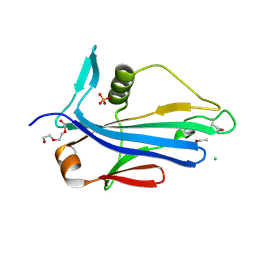 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3K1S
 
 | | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PTS system, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the PTS Cellobiose Specific Enzyme IIA from Bacillus anthracis
To be Published
|
|
3S42
 
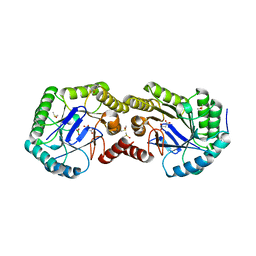 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site | | Descriptor: | 3-dehydroquinate dehydratase, BORIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Halavaty, A.S, Krishna, S.N, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site
To be Published
|
|
5CXD
 
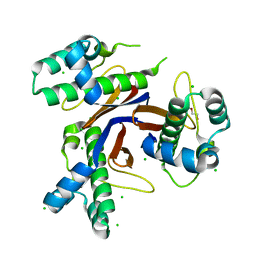 | | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Holo-[acyl-carrier-protein] synthase, ... | | Authors: | Halavaty, A.S, Minasov, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
To Be Published
|
|
1TT4
 
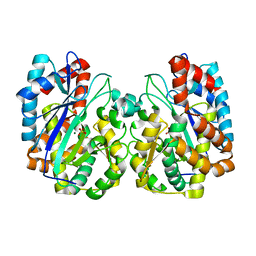 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
3SEF
 
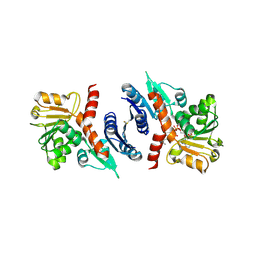 | | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate 5-dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH
To be Published
|
|
3K96
 
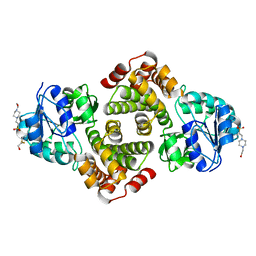 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
2I0Z
 
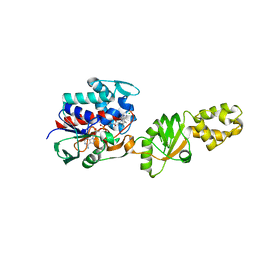 | | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(FAD)-utilizing dehydrogenases | | Authors: | Minasov, G, Shuvalova, L, Vorontsov, I.I, Kiryukhina, O, Abdullah, J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of a FAD binding protein from Bacillus cereus, a putative NAD(FAD)-utilizing dehydrogenases
To be Published
|
|
4ZXU
 
 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
2W27
 
 | | CRYSTAL STRUCTURE OF THE BACILLUS SUBTILIS YKUI PROTEIN, WITH AN EAL DOMAIN, IN COMPLEX WITH SUBSTRATE C-DI-GMP AND CALCIUM | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, YKUI PROTEIN | | Authors: | Padavattan, S, AndERSON, W.F, Schirmer, T. | | Deposit date: | 2008-10-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ykui and its Complex with Second Messenger C-Di-Gmp Suggests Catalytic Mechanism of Phosphodiester Bond Cleavage by Eal Domains.
J.Biol.Chem., 284, 2009
|
|
3SLH
 
 | | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 1,2-ETHANEDIOL, 3-phosphoshikimate 1-carboxyvinyltransferase, ... | | Authors: | Light, S.H, Minasov, G, Filippova, E.V, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase(AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate
To be Published
|
|
4YO4
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment phthalazine | | Descriptor: | ACETATE ION, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
4YPD
 
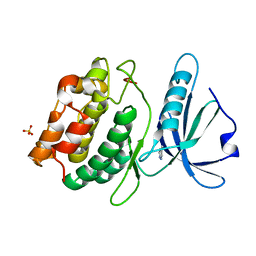 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
5JRO
 
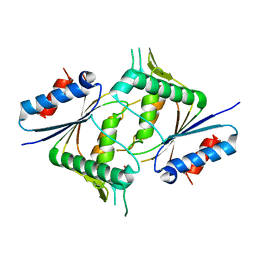 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
5JPD
 
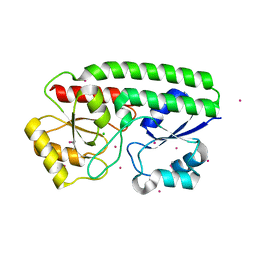 | | Metal ABC transporter from Listeria monocytogenes with cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with cadmium
to be published
|
|
5CRF
 
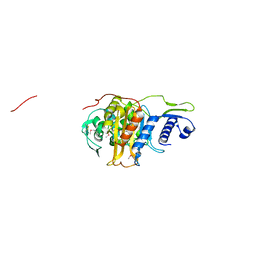 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5DGX
 
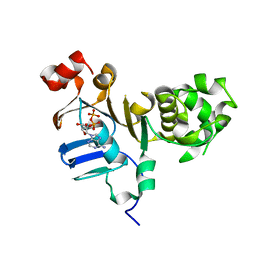 | | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
5D6A
 
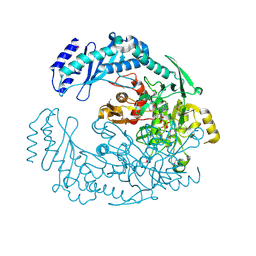 | | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Predicted ATPase of the ABC class, SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP)
To Be Published
|
|
5CXW
 
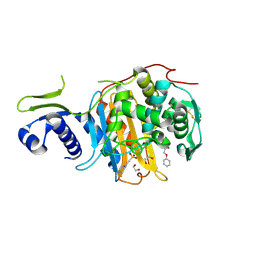 | | Structure of the PonA1 protein from Mycobacterium Tuberculosis in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5DJ9
 
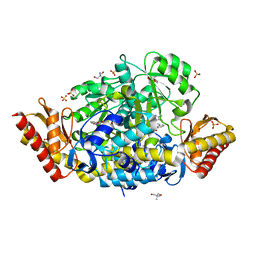 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine
To Be Published
|
|
5CQE
 
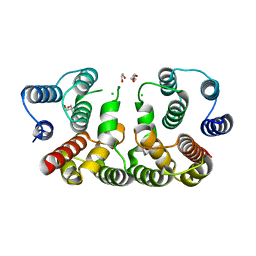 | | 2.1 Angstrom resolution crystal structure of matrix protein 1 (M1; residues 1-164) from Influenza A virus (A/Puerto Rico/8/34(H1N1)) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Flores, K, Dubrovska, I, Grimshaw, S, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of matrix protein 1 (M1; residues 1-164) from Influenza A virus (A/Puerto Rico/8/34(H1N1))
To Be Published
|
|
4ZND
 
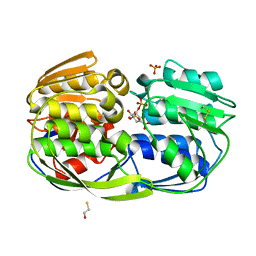 | | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL, PHOSPHATE ION, ... | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium
To Be Published
|
|
4ZWL
 
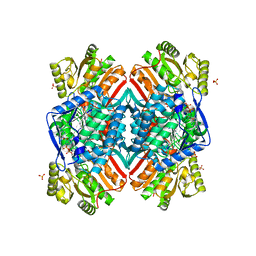 | | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-19 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.60 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289
To be Published
|
|
3UPD
 
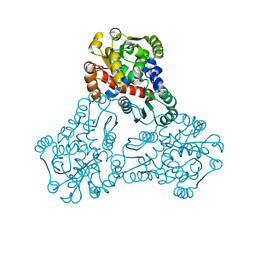 | | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus.
TO BE PUBLISHED
|
|
