6TNI
 
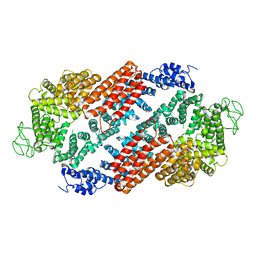 | | Structure of FANCD2 homodimer | | Descriptor: | Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
9FFF
 
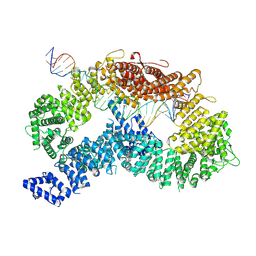 | | dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (32-MER), DNA (33-MER), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-23 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
9FFB
 
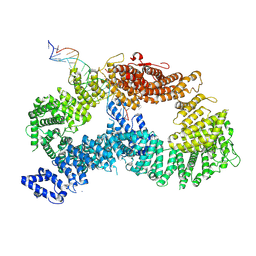 | | ss-dsDNA-FANCD2-FANCI complex | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*GP*TP*CP*TP*CP*TP*AP*GP*AP*CP*AP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*GP*CP*TP*GP*TP*CP*TP*AP*GP*AP*GP*AP*CP*AP*TP*CP*GP*AP*T)-3'), Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Passmore, L.A. | | Deposit date: | 2024-05-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | FANCD2-FANCI surveys DNA and recognizes double- to single-stranded junctions.
Nature, 632, 2024
|
|
6TNG
 
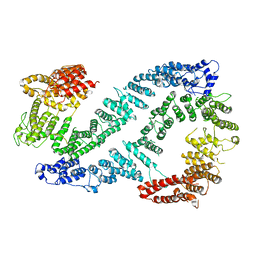 | | Structure of FANCD2 in complex with FANCI | | Descriptor: | Fanconi anemia complementation group I, Uncharacterized protein | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6TNF
 
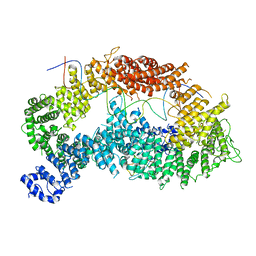 | | Structure of monoubiquitinated FANCD2 in complex with FANCI and DNA | | Descriptor: | DNA (33-MER), FANCD2, Fanconi anemia complementation group I, ... | | Authors: | Alcon, P, Shakeel, S, Passmore, L.A. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | FANCD2-FANCI is a clamp stabilized on DNA by monoubiquitination of FANCD2 during DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8A2Q
 
 | |
7AAL
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1, G258A mutant | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAM
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 bound to the CTH domain of the phosphatase LYP | | Descriptor: | GLYCEROL, Proline-serine-threonine phosphatase-interacting protein 1, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAN
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
6SRI
 
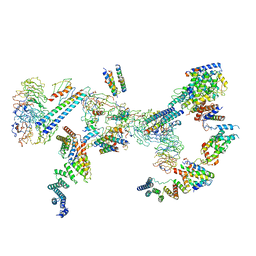 | | Structure of the Fanconi anaemia core complex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (base region, proposed FANCC-FANC-E-FANCF), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
6SRS
 
 | | Structure of the Fanconi anaemia core subcomplex | | Descriptor: | Fanconi anaemia protein FANCL, Unassigned secondary structure elements (central region, proposed FANCB-FAAP100), ... | | Authors: | Shakeel, S, Rajendra, E, Alcon, P, He, S, Scheres, S.H.W, Passmore, L.A. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Fanconi anaemia monoubiquitin ligase complex.
Nature, 575, 2019
|
|
5MGA
 
 | |
6GTC
 
 | | Transition state structure of Cpf1(Cas12a) I1 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Mesa, P, Montoya, G. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTE
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I3 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTG
 
 | | Transition state structure of Cpf1(Cas12a) I4 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (32-MER), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), ... | | Authors: | Mesa, P, Montoya, G, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTD
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I2 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*AP*GP*T)-3'), DNA (5'-D(P*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), ... | | Authors: | Montoya, G, Mesa, P. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.24 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
6GTF
 
 | | Transient state structure of CRISPR-Cpf1 (Cas12a) I5 conformation | | Descriptor: | CRISPR-associated endonuclease Cas12a, DNA (5'-D(P*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*TP*AP*AP*CP*AP*AP*GP*CP*TP*CP*G)-3'), DNA (5'-D(P*CP*GP*AP*GP*CP*TP*CP*GP*TP*TP*AP*GP*AP*GP*AP*A)-3'), ... | | Authors: | Montoya, G, Mesa, P, Stella, S. | | Deposit date: | 2018-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity.
Cell, 175, 2018
|
|
