7DGF
 
 | |
7DGG
 
 | |
7DGI
 
 | |
8TQG
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20419 | | Descriptor: | N-benzyl-2-{4-[4-(4,5-dimethoxy-1H-indole-2-carbonyl)piperazine-1-carbonyl]piperidin-1-yl}-6-methylpyrimidine-4-carboxamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TR4
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20404 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N,N-dimethylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TRY
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20348 | | Descriptor: | N-{(2S,3S)-4-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-3-hydroxy-1-phenylbutan-2-yl}-4-(2-methylbutan-2-yl)benzene-1-sulfonamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-10 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
6XD9
 
 | | Carbonmonoxy hemoglobin in complex with the antisickling agent 2-hydroxy-6-((6-(hydroxymethyl)pyridin-2-yl)methoxy)benzaldehyde (VZHE039) | | Descriptor: | 3-{[6-(hydroxymethyl)pyridin-2-yl]methoxy}-2-methylphenol, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Pagare, P.P, Safo, M.K, Musayev, F.N. | | Deposit date: | 2020-06-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | VZHE-039, a novel antisickling agent that prevents erythrocyte sickling under both hypoxic and anoxic conditions.
Sci Rep, 10, 2020
|
|
6XDT
 
 | | Carbonmonoxy hemoglobin in complex with the antisickling agent methyl 5-((2-formyl-4-methoxyphenoxy)methyl)picolinate | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Pagare, P.P, Safo, M.K, Musayev, F.N. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploration of Structure-Activity Relationship of Aromatic Aldehydes Bearing Pyridinylmethoxy-Methyl Esters as Novel Antisickling Agents.
J.Med.Chem., 63, 2020
|
|
6WJ0
 
 | | Crystal structure of Fab 54-4H03 | | Descriptor: | Fab 54-4H03 heavy chain, Fab 54-4H03 light chain, GLYCEROL | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
6WJ1
 
 | | Crystal structure of Fab 54-4H03 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-4H03 heavy chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
7AVY
 
 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVX
 
 | | MerTK kinase domain in complex with NPS-1034 | | Descriptor: | 1-(4-fluorophenyl)-N-[3-fluoro-4-[(3-phenyl-1H-pyrrolo[2,3-b]pyridin-4-yl)oxy]phenyl]-2,3-dimethyl-5-oxopyrazole-4-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW4
 
 | | MerTK kinase domain with type 3 inhibitor from a DNA-encoded library | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-(4-methyl-1,2,3-thiadiazol-5-yl)-N-((R)-1-(((R)-3-(methylamino)-3-oxo-1-(4-(trifluoromethyl)phenyl)propyl)amino)-1-oxo-4-phenylbutan-2-yl)isoxazole-4-carboxamide, CHLORIDE ION, ... | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW1
 
 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AVZ
 
 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW3
 
 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
3OEC
 
 | |
6WW4
 
 | | Crystal structure of HERC2 ZZ domain in complex with histone H3 tail | | Descriptor: | GLYCEROL, Histone H3.1,E3 ubiquitin-protein ligase HERC2, ZINC ION | | Authors: | Liu, J, Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-07 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural Insight into Binding of the ZZ Domain of HERC2 to Histone H3 and SUMO1.
Structure, 28, 2020
|
|
6LNQ
 
 | | The co-crystal structure of SARS-CoV 3C Like Protease with aldehyde inhibitor M7 | | Descriptor: | N-[(2S)-3-methyl-1-[[(2S)-4-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]amino]-1-oxidanylidene-butan-2-yl]-1H-indole-2-carboxamide, Severe Acute Respiratory Syndrome Coronavirus 3c Like Protease | | Authors: | Wang, H, Shang, L.Q. | | Deposit date: | 2020-01-01 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Comprehensive Insights into the Catalytic Mechanism of Middle East Respiratory Syndrome 3C-Like Protease and Severe Acute Respiratory Syndrome 3C-Like Protease.
Acs Catalysis, 10, 2020
|
|
6LO0
 
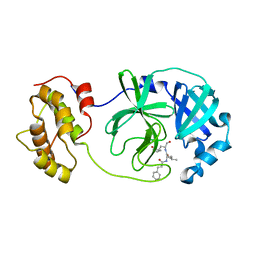 | |
7WKS
 
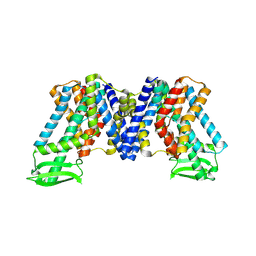 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
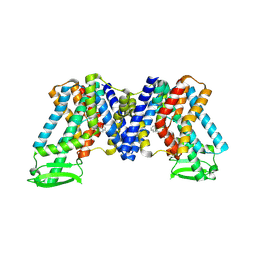 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
3FMZ
 
 | |
5T18
 
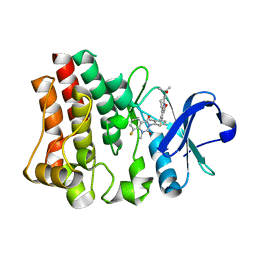 | | Crystal structure of Bruton agammabulinemia tyrosine kinase complexed with BMS-986142 aka (2s)-6-fluoro-5-[3-(8-fluoro-1-methyl-2,4-dioxo-1,2,3,4-tetrahydroquinazolin-3-yl)-2-methylphenyl]-2-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1h-carbazole-8-carboxamide | | Descriptor: | 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-08-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 6-Fluoro-5-(R)-(3-(S)-(8-fluoro-1-methyl-2,4-dioxo-1,2-dihydroquinazolin-3(4H)-yl)-2-methylphenyl)-2-(S)-(2-hydroxypropan-2-yl)-2,3,4,9-tetrahydro-1H-carbazole-8-carboxamide (BMS-986142): A Reversible Inhibitor of Bruton's Tyrosine Kinase (BTK) Conformationally Constrained by Two Locked Atropisomers.
J. Med. Chem., 59, 2016
|
|
