6U9R
 
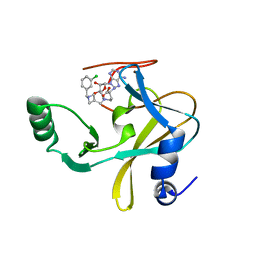 | | MLL1 SET N3861I/Q3867L bound to inhibitor 12 (TC-5140) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(3-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
2GQB
 
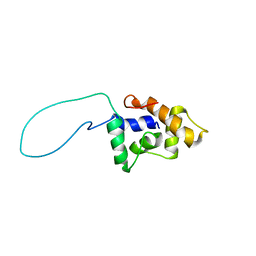 | | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris; (Northeast Structural Genomics Consortium Target RpT4; Ontario Centre for Structural Proteomics Target rp2812 ) | | Descriptor: | conserved hypothetical protein | | Authors: | Srisailam, S, Lukin, J.A, Yee, A, Lemak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-20 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a conserved unknown protein RPA2825 from Rhodopseudomonas palustris
To be Published
|
|
2H71
 
 | |
2GPF
 
 | |
2H4Y
 
 | |
6U64
 
 | | Mcl-1 bound to compound 17 | | Descriptor: | 5-[(2-phenylethyl)sulfanyl]-2-{[(4-phenylpiperazin-1-yl)sulfonyl]amino}benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6U67
 
 | | Mcl-1 bound to compound 24 | | Descriptor: | 2-({[4-(4-tert-butylphenyl)piperazin-1-yl]sulfonyl}amino)-5-{[3-oxo-3-(phenylamino)propyl]sulfanyl}benzoic acid, BIPHENYL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6ROD
 
 | |
6ROL
 
 | | Structure of IMP2 KH34 domains | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Insulin-like growth factor 2 mRNA-binding protein 2, ... | | Authors: | Bhaskar, V, Biswas, J, Singer, R.H, Chao, J.A. | | Deposit date: | 2019-05-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for RNA selectivity by the IMP family of RNA-binding proteins.
Nat Commun, 10, 2019
|
|
2KYH
 
 | |
6S3Y
 
 | |
6ROH
 
 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
2HFC
 
 | | Structure of S65T Y66F R96A GFP variant in precursor state | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Barondeau, D.P, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2006-06-23 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Case of the Missing Ring: Radical Cleavage of a Carbon-Carbon Bond and Implications for GFP Chromophore Biosynthesis
J.Am.Chem.Soc., 129, 2007
|
|
6S37
 
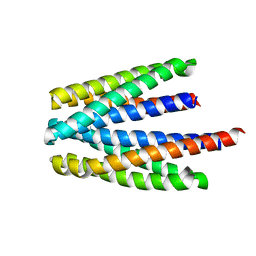 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
2H76
 
 | |
2H7J
 
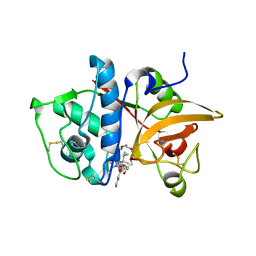 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor. | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
2H75
 
 | |
6TY6
 
 | | Variant W229D/F290W-2 of the last common ancestor of Gram-negative bacteria beta-lactamase class A (GNCA4) bound to 5(6)-nitrobenzotriazole (TS-analog) | | Descriptor: | 6-NITROBENZOTRIAZOLE, ACETATE ION, Beta lactamase (GNCA4-2), ... | | Authors: | Gavira, J.A, Risso, V, Sanchez-Ruiz, J.M, Romero-Rivera, A, Kamerlin, S.C.L, Ortega-Munoz, M, Santoyo-Gonzalez, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enhancing ade novoenzyme activity by computationally-focused ultra-low-throughput screening.
Chem Sci, 11, 2020
|
|
2GVG
 
 | | Crystal Structure of human NMPRTase and its complex with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Khan, J.A, Tao, X, Tong, L. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
Nat.Struct.Mol.Biol., 13, 2006
|
|
6U9M
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U8D
 
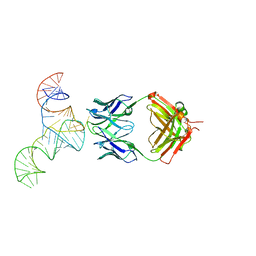 | | Crystal structure of hepatitis C virus IRES junction IIIabc in complex with Fab HCV2 | | Descriptor: | Heavy chain of Fab HCV2, JIIIabc RNA (68-MER), Light chain of Fab HCV2 | | Authors: | Koirala, D, Lewicka, A, Koldobskaya, Y, Huang, H, Piccirilli, J.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Synthetic Antibody Binding to a Preorganized RNA Domain of Hepatitis C Virus Internal Ribosome Entry Site Inhibits Translation.
Acs Chem.Biol., 15, 2020
|
|
2KUB
 
 | | Solution structure of the alpha subdomain of the major non-repeat unit of Fap1 fimbriae of Streptococcus parasanguis | | Descriptor: | Fimbriae-associated protein Fap1 | | Authors: | Ramboarina, S, Garnett, J.A, Bodey, A, Simpson, P, Bardiaux, B, Nilges, M, Matthews, S. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into serine-rich fimbriae from gram-positive bacteria.
J.Biol.Chem., 2010
|
|
2HXZ
 
 | | Crystal Structure of Cathepsin S in complex with a Nonpeptidic Inhibitor (Hexagonal spacegroup) | | Descriptor: | Cathepsin S, N-[(1S)-1-{1-[(1R,3E)-1-ACETYLPENT-3-EN-1-YL]-1H-1,2,3-TRIAZOL-4-YL}-1,2-DIMETHYLPROPYL]BENZAMIDE, SULFATE ION | | Authors: | Patterson, A.W, Wood, W.J, Hornsby, M, Lesley, S, Spraggon, G, Ellman, J.A. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of selective, nonpeptidic nitrile inhibitors of cathepsin s using the substrate activity screening method.
J.Med.Chem., 49, 2006
|
|
2I5B
 
 | | The crystal structure of an ADP complex of Bacillus subtilis pyridoxal kinase provides evidence for the parralel emergence of enzyme activity during evolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphomethylpyrimidine kinase | | Authors: | Newman, J.A, Das, S.K, Sedelnikova, S.E, Rice, D.W. | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of an ADP Complex of Bacillus subtilis Pyridoxal Kinase Provides Evidence for the Parallel Emergence of Enzyme Activity During Evolution.
J.Mol.Biol., 363, 2006
|
|
2H51
 
 | |
