8SJP
 
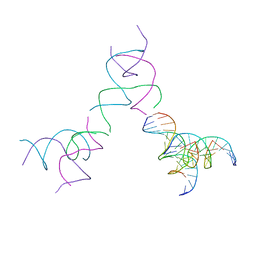 | | [3T15] Self-assembling DNA motif with 15 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*TP*GP*AP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*CP*TP*GP*TP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*CP*GP*AP*TP*GP*GP*AP*CP*AP*GP*GP*GP*G)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (5.22 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJN
 
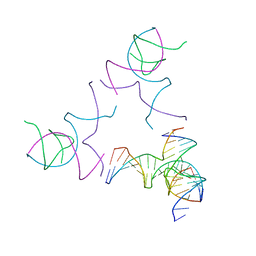 | | [3T13] Self-assembling left-handed tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*TP*CP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*TP*GP*TP*GP*GP*CP*GP*AP*TP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.3 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJU
 
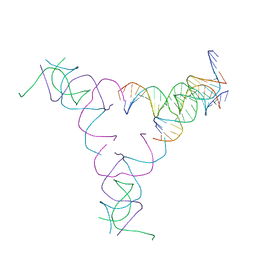 | | [4T17] Self-assembling right-handed four-turn tensegrity triangle with 17 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (25-MER), DNA (5'-D(*GP*AP*AP*AP*AP*AP*CP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*GP*GP*TP*AP*TP*TP*CP*AP*CP*CP*AP*CP*GP*AP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.14 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJS
 
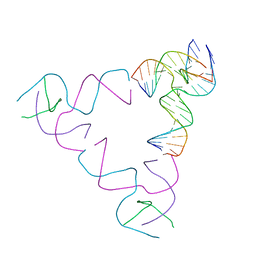 | | [3T18] Self-assembling right-handed tensegrity triangle with 18 interjunction base pairs and P63 symmetry | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*GP*CP*CP*TP*GP*AP*CP*AP*TP*AP*CP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*CP*TP*GP*TP*GP*GP*CP*TP*C)-3'), DNA (5'-D(P*TP*CP*GP*TP*GP*GP*AP*CP*AP*GP*CP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.31 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJW
 
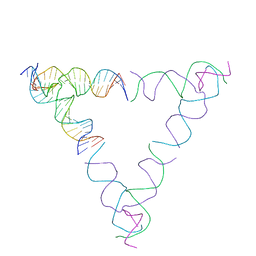 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.64 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJO
 
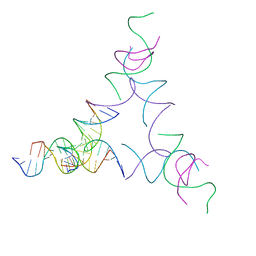 | | [3T14] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(P*CP*AP*CP*GP*TP*GP*GP*AP*CP*AP*GP*GP*AP*G)-3'), DNA (5'-D(P*CP*AP*GP*CP*TP*CP*AP*GP*CP*CP*TP*GP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*GP*TP*GP*AP*GP*TP*CP*TP*CP*CP*AP*CP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.08 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SJT
 
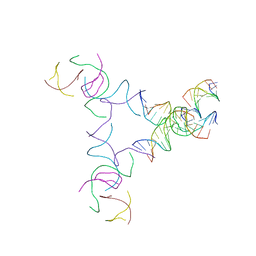 | | [3T14+10] Self-assembling left-handed tensegrity triangle with 14 interjunction base pairs and a 10 bp linker with R3 symmetry | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*CP*CP*TP*GP*AP*GP*GP*TP*CP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*AP*CP*TP*CP*TP*GP*CP*TP*A)-3'), DNA (5'-D(*GP*TP*TP*AP*GP*CP*AP*GP*AP*G)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (9.38 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SL5
 
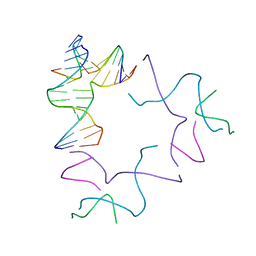 | | [2T13] Self-assembling left-handed two-turn tensegrity triangle with 13 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*GP*AP*CP*TP*AP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*TP*GP*GP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*AP*CP*A)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.55 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8SZ5
 
 | | [2T5] Self-assembling DNA motif with 5 base pairs between junctions and P32 symmetry | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(P*AP*CP*GP*AP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*GP*T)-3'), ... | | Authors: | Vecchioni, S, Janowski, J, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-05-26 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4CP8
 
 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | Descriptor: | ALLOPHANATE HYDROLASE, MALONATE ION | | Authors: | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | Deposit date: | 2014-02-03 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
5EAK
 
 | |
2QHN
 
 | | Crystal Structure of Chek1 in Complex with Inhibitor 1a | | Descriptor: | 5-ETHYL-3-METHYL-1,5-DIHYDRO-4H-PYRAZOLO[4,3-C]QUINOLIN-4-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2QHM
 
 | | crystal structure of Chek1 in complex with inhibitor 2a | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-PYRROLO[2,3-B]PYRIDINE-3-CARBOXYLATE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S. | | Deposit date: | 2007-07-02 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Optimization of a pyrazoloquinolinone class of Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4OON
 
 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
2HXL
 
 | | crystal structure of Chek1 in complex with inhibitor 1 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)-1H-INDAZOLE-6-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4OOL
 
 | | Crystal structure of PBP3 in complex with compound 14 ((2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid) | | Descriptor: | (2E)-2-({[(2S)-2-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}acetyl]amino}-3-oxopropyl]oxy}imino)pentanedioic acid, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
2HXQ
 
 | | crystal structure of Chek1 in complex with inhibitor 2 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)QUINOLIN-2(1H)-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HY0
 
 | | crystal structure of chek1 in complex with inhibitor 22 | | Descriptor: | 3-[5-(PIPERIDIN-1-YLMETHYL)-1H-INDOL-2-YL]-6-(1H-PYRAZOL-4-YL)QUINOLIN-2(1H)-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4OOM
 
 | | Crystal structure of PBP3 in complex with BAL30072 ((2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide) | | Descriptor: | (2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)methoxy]imino}-N-{(2S)-1-hydroxy-3-methyl-3-[(sulfooxy)amino]butan-2-yl}ethanamide, Cell division protein FtsI [Peptidoglycan synthetase] | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2014-05-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
2SPZ
 
 | | STAPHYLOCOCCAL PROTEIN A, Z-DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN A | | Authors: | Montelione, G.T, Tashiro, M, Tejero, R, Lyons, B.A. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution NMR structure of the Z domain of staphylococcal protein A.
J.Mol.Biol., 272, 1997
|
|
1QP9
 
 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
3F9N
 
 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | Descriptor: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S, Ikuta, M. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1HCV
 
 | |
2VCP
 
 | | Crystal structure of N-Wasp VC domain in complex with skeletal actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gaucher, J.F, Didry, D, Carlier, M.F. | | Deposit date: | 2007-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex.
J. Biol. Chem., 287, 2012
|
|
2HOG
 
 | | crystal structure of Chek1 in complex with inhibitor 20 | | Descriptor: | (5-{3-[5-(PIPERIDIN-1-YLMETHYL)-1H-INDOL-2-YL]-1H-INDAZOL-6-YL}-2H-1,2,3-TRIAZOL-4-YL)METHANOL, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Ikuta, M. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-(Indol-2-yl)indazoles as Chek1 kinase inhibitors: Optimization of potency and selectivity via substitution at C6.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
