8HM1
 
 | |
8HM3
 
 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
7YK4
 
 | | ox40-antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor receptor superfamily member 4, antibody-H, ... | | Authors: | Zhou, A. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of a Novel Agonistic Anti-OX40 Antibody.
Biomolecules, 12, 2022
|
|
7C20
 
 | |
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 98, 2024
|
|
6VHH
 
 | | Human Teneurin-2 and human Latrophilin-3 binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L3, ... | | Authors: | Xie, Y, Li, J, Arac, D, Zhao, M. | | Deposit date: | 2020-01-09 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism.
Nat Commun, 11, 2020
|
|
5H19
 
 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
6IS5
 
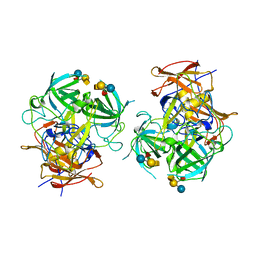 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IR5
 
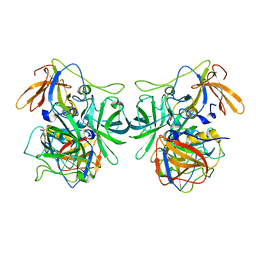 | | P domain of GII.3-TV24 | | Descriptor: | VP1 Capsid protein | | Authors: | Yang, Y. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6J0Q
 
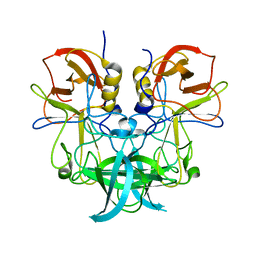 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
3BY1
 
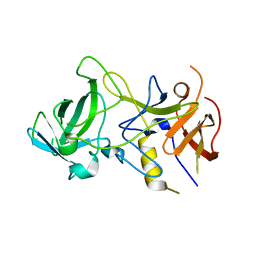 | | Unliganded Norvalk Virus P domain | | Descriptor: | 58 kd capsid protein | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-01-15 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
6M2B
 
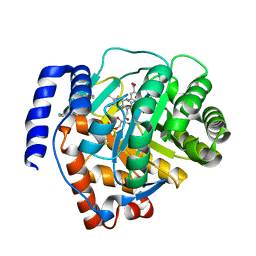 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H. | | Deposit date: | 2020-02-27 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2.
Protein Cell, 11, 2020
|
|
4ZWB
 
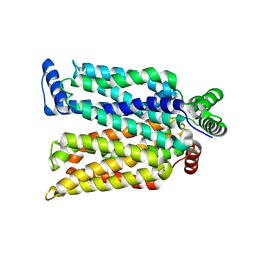 | | Crystal structure of maltose-bound human GLUT3 in the outward-occluded conformation at 2.4 angstrom | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 3, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZWC
 
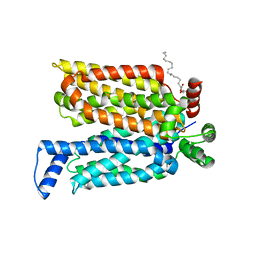 | | Crystal structure of maltose-bound human GLUT3 in the outward-open conformation at 2.6 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
4ZW9
 
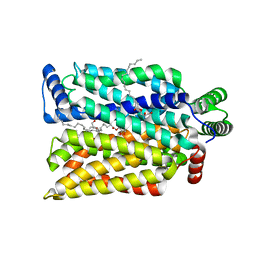 | | Crystal structure of human GLUT3 bound to D-glucose in the outward-occluded conformation at 1.5 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Solute carrier family 2, facilitated glucose transporter member 3, ... | | Authors: | Deng, D, Sun, P.C, Yan, C.Y, Yan, N. | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Molecular basis of ligand recognition and transport by glucose transporters
Nature, 526, 2015
|
|
5L4W
 
 | | Crystal structure of FimH lectin domain in complex with 3-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
3D26
 
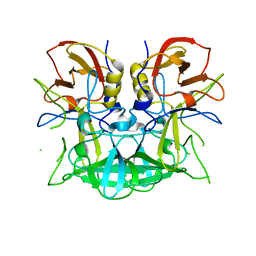 | | Norwalk P domain A-trisaccharide complex | | Descriptor: | 58 kd capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
5L4X
 
 | | Crystal structure of FimH lectin domain in complex with 4-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4U
 
 | | Crystal structure of FimH lectin domain in complex with 2-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4Y
 
 | | Crystal structure of FimH lectin domain in complex with 4-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4V
 
 | | Crystal structure of FimH lectin domain in complex with 3-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4T
 
 | | Crystal structure of FimH lectin domain in complex with 2-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
8XIF
 
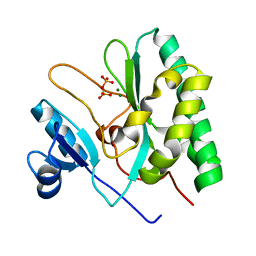 | | The crystal structure of the AEP domain of VACV D5 | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, Uncoating factor OPG117 | | Authors: | Gan, J, Zhang, W. | | Deposit date: | 2023-12-19 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ8
 
 | | The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
