5NVB
 
 | |
5N5B
 
 | |
5N5A
 
 | |
1LCJ
 
 | |
1LCK
 
 | |
6Z1E
 
 | |
6Z1D
 
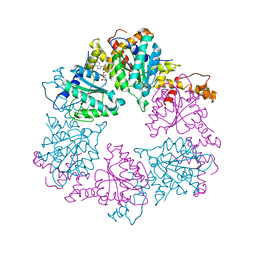 | |
3NI0
 
 | | Crystal Structure of Mouse BST-2/Tetherin Ectodomain | | Descriptor: | Bone marrow stromal antigen 2, ISOPROPYL ALCOHOL | | Authors: | Scheaffer, S.M, Brett, T.J. | | Deposit date: | 2010-06-14 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biophysical Analysis of BST-2/Tetherin Ectodomains Reveals an Evolutionary Conserved Design to Inhibit Virus Release.
J.Biol.Chem., 286, 2011
|
|
7ZWG
 
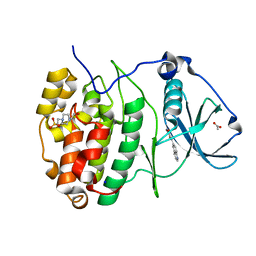 | | The Crystal structure of RO4493940 bound to CK2alpha | | Descriptor: | (5~{Z})-5-(quinolin-6-ylmethylidene)-1,3-thiazolidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
7ZWE
 
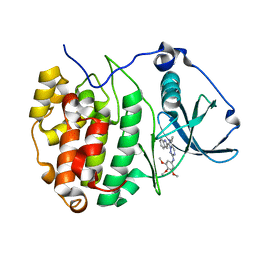 | | The Crystal structure of GW8695 bound to CK2alpha | | Descriptor: | 7-(1~{H}-indol-2-yl)-5-methyl-~{N}-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 20, 2024
|
|
7A4Q
 
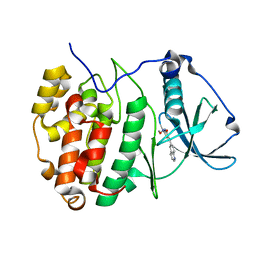 | | The Crystal structure of RO4613269 bound to CK2alpha | | Descriptor: | 2-methoxyimino-5-(quinolin-6-ylmethyl)-1,3-thiazol-4-one, Casein kinase II subunit alpha | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-08-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Nat.Chem.Biol., 2023
|
|
6HAS
 
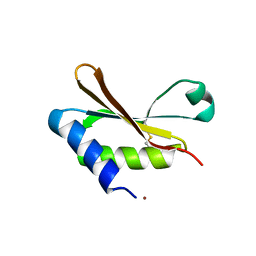 | |
6FYH
 
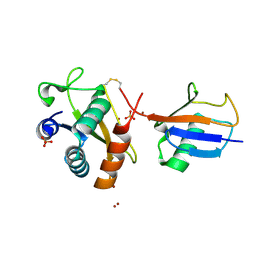 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
6FX4
 
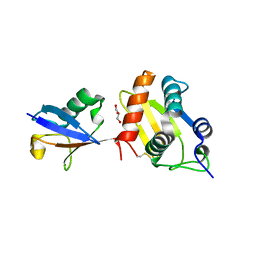 | |
1NKF
 
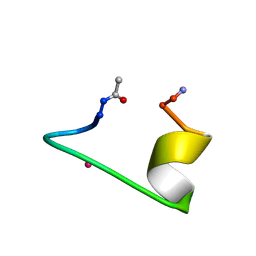 | | CALCIUM-BINDING PEPTIDE, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM-BINDING HEXADECAPEPTIDE, LANTHANUM (III) ION | | Authors: | Sticht, H, Ejchart, A. | | Deposit date: | 1998-03-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Alpha-helix nucleation by a calcium-binding peptide loop.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3R7G
 
 | | Crystal structure of Spire KIND domain in complex with the tail of FMN2 | | Descriptor: | Formin-2, Protein spire homolog 1 | | Authors: | Kreutz, B, Vizcarra, C.L, Rodal, A.A, Toms, A.V, Lu, J, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
3RBW
 
 | | Crystal structure of Spire KIND domain | | Descriptor: | Protein spire homolog 1 | | Authors: | Vizcarra, C.L, Kreutz, B, Rodal, A.A, Toms, A.V, Lu, J, Zheng, W, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-30 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
7NKT
 
 | | RBD domain of SARS-CoV2 in complex with neutralizing nanobody NM1226 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Ostertag, E, Zocher, G, Stehle, T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Embo Rep., 22, 2021
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
6RXK
 
 | | Crystal structure of CobB wt in complex with H4K16-Butyryl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXL
 
 | | Crystal structure of CobB wt in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXR
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162G) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after co-crystallisation | | Descriptor: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXO
 
 | | Crystal structure of CobB Ac2 (A76G, I131C, V162A) in complex with H4K16-Buturyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXP
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16-Crotonyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
