4I8Y
 
 | | Structure of the unliganded N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I90
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to choline | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-04 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9M
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus bound to HEPES | | Descriptor: | 1-phosphatidylinositol phosphodiesterase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9T
 
 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
7XNX
 
 | |
7XNY
 
 | |
8JTV
 
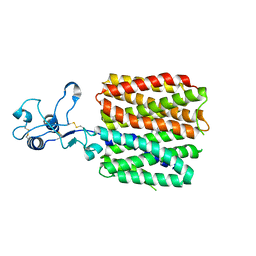 | | hOCT1 in complex with metformin in inward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
3C6R
 
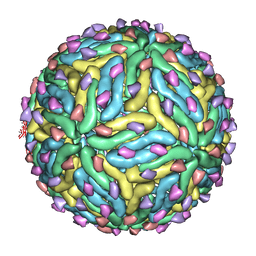 | | Low pH Immature Dengue Virus | | Descriptor: | Envelope protein, Peptide pr | | Authors: | Yu, I, Zhang, W, Holdway, H.A, Li, L, Kostyuchenko, V.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G, Chen, J. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Structure of the immature dengue virus at low pH primes proteolytic maturation
Science, 319, 2008
|
|
6X4Y
 
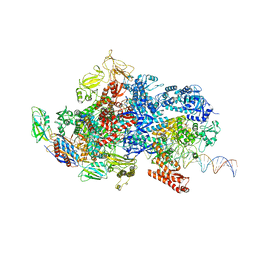 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X50
 
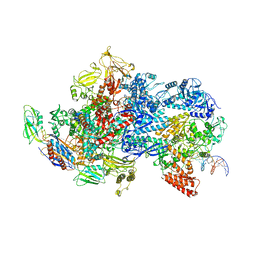 | | Mfd-bound E.coli RNA polymerase elongation complex - V state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewelyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X26
 
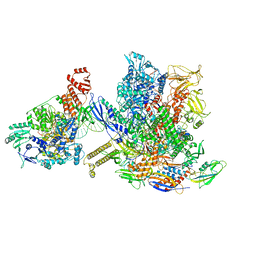 | | Mfd-bound E.coli RNA polymerase elongation complex - L1 state | | Descriptor: | DNA (64-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X43
 
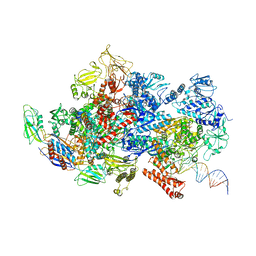 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
1M4M
 
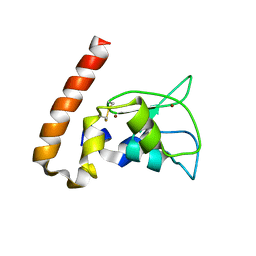 | | Mouse Survivin | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 5, ZINC ION | | Authors: | Muchmore, S.W, Chen, J, Jakob, C, Zakula, D, Matayoshi, E.D, Wu, W, Zhang, H, Li, F, Ng, S.C, Altieri, D.C. | | Deposit date: | 2002-07-03 | | Release date: | 2002-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE AND MUTAGENIC ANALYSIS OF THE INHIBITOR-OF-APOPTOSIS PROTEIN SURVIVIN
MOL.CELL, 6, 2000
|
|
7YTJ
 
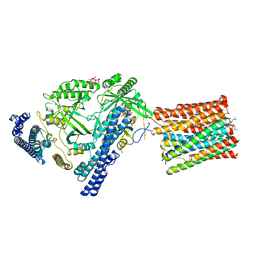 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
7DL2
 
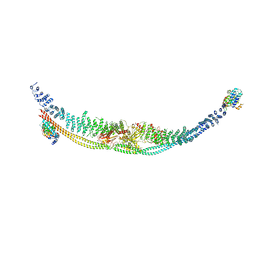 | | Cryo-EM structure of human TSC complex | | Descriptor: | Hamartin, Isoform 7 of Tuberin, TBC1 domain family member 7, ... | | Authors: | Yang, H, Yu, Z, Chen, X, Li, J, Li, N, Cheng, J, Gao, N, Yuan, H, Ye, D, Guan, K, Xu, Y. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into TSC complex assembly and GAP activity on Rheb.
Nat Commun, 12, 2021
|
|
6L6W
 
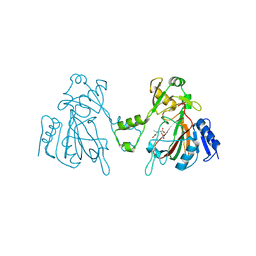 | | The structure of ScoE with intermediate | | Descriptor: | (3R)-3-[[(1R)-1,2-bis(oxidanyl)-2-oxidanylidene-ethyl]amino]butanoic acid, FE (II) ION, FORMIC ACID, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6TNU
 
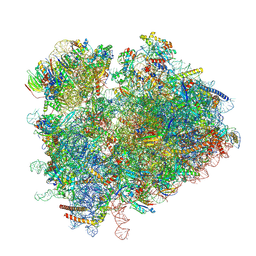 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
1Q09
 
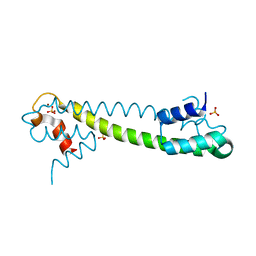 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q0A
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group C222) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q06
 
 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | SILVER ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q08
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator, at 1.9 A resolution (space group P212121) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q05
 
 | | Crystal structure of the Cu(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | COPPER (I) ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q07
 
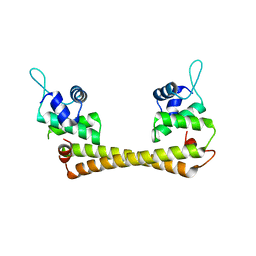 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
7TZO
 
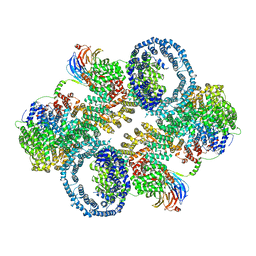 | | The apo structure of human mTORC2 complex | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Yu, Z, Chen, J, Pearce, D. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Interactions between mTORC2 core subunits Rictor and mSin1 dictate selective and context-dependent phosphorylation of substrate kinases SGK1 and Akt.
J.Biol.Chem., 298, 2022
|
|
