2XYK
 
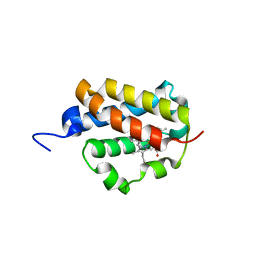 | | Group II 2-on-2 Hemoglobin from the Plant Pathogen Agrobacterium tumefaciens | | Descriptor: | 2-ON-2 HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Nardini, M, LaBarre, M, Richard, C, Wittenberg, J.B, Wittenberg, B.A, Guertin, M, Bolognesi, M. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of a Group II 2/2 Hemoglobin from the Plant Pathogen Agrobacterium Tumefaciens.
Biochim.Biophys.Acta, 1814, 2011
|
|
1G7E
 
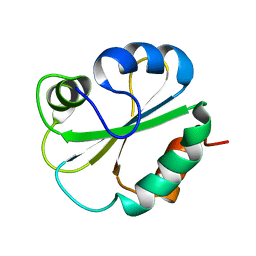 | | NMR STRUCTURE OF N-DOMAIN OF ERP29 PROTEIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, M, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1S9H
 
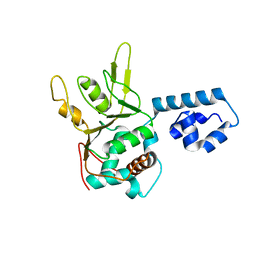 | | Crystal Structure of Adeno-associated virus Type 2 Rep40 | | Descriptor: | Rep 40 protein | | Authors: | James, J.A, Escalante, C.R, Yoon-Robarts, M, Edwards, T.A, Linden, R.M, Aggarwal, A.K. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the SF3 Helicase from Adeno-Associated Virus Type 2
Structure, 11, 2003
|
|
3IWF
 
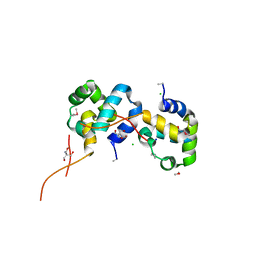 | | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-METHOXYETHANOL, CHLORIDE ION, ... | | Authors: | Stein, A.J, Sather, A, Borovilos, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the N-terminal domain of a RpiR Transcriptional Regulator from Staphylococcus epidermidis to 1.4A
To be Published
|
|
1SKY
 
 | | CRYSTAL STRUCTURE OF THE NUCLEOTIDE FREE ALPHA3BETA3 SUB-COMPLEX OF F1-ATPASE FROM THE THERMOPHILIC BACILLUS PS3 | | Descriptor: | F1-ATPASE, SULFATE ION | | Authors: | Shirakihara, Y, Leslie, A.G.W, Abrahams, J.P, Walker, J.E, Ueda, T, Sekimoto, Y, Kambara, M, Saika, K, Kagawa, Y, Yoshida, M. | | Deposit date: | 1997-02-26 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of the nucleotide-free alpha 3 beta 3 subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer.
Structure, 5, 1997
|
|
2WBM
 
 | | Crystal structure of mthSBDS, the homologue of the Shwachman-Bodian- Diamond syndrome protein in the euriarchaeon Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, GLYCEROL, RIBOSOME MATURATION PROTEIN SDO1 HOMOLOG, ... | | Authors: | Ng, C.L, Isupov, M.N, Lebedev, A.A, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Flexibility and Molecular Interactions of an Archaeal Homologue of the Shwachman-Bodian-Diamond Syndrome Protein.
Bmc Struct.Biol., 9, 2009
|
|
2R2Z
 
 | | The crystal structure of a hemolysin domain from Enterococcus faecalis V583 | | Descriptor: | Hemolysin, ZINC ION | | Authors: | Zhang, R, Tan, K, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of a hemolysin domain from Enterococcus faecalis V583.
To be Published
|
|
2Q89
 
 | | Crystal structure of EhuB in complex with hydroxyectoine | | Descriptor: | (4S,5S)-5-HYDROXY-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
3CHG
 
 | | The compatible solute-binding protein OpuAC from Bacillus subtilis in complex with DMSA | | Descriptor: | (dimethyl-lambda~4~-sulfanyl)acetic acid, Glycine betaine-binding protein | | Authors: | Smits, S.H.J, Hoing, M, Lecher, J, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2008-03-09 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Compatible-Solute-Binding Protein OpuAC from Bacillus subtilis: Ligand Binding, Site-Directed Mutagenesis, and Crystallographic Studies
J.Bacteriol., 190, 2008
|
|
3CZX
 
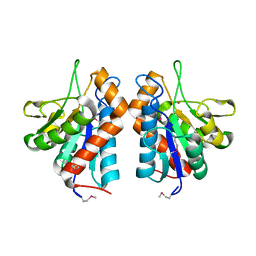 | |
2PJQ
 
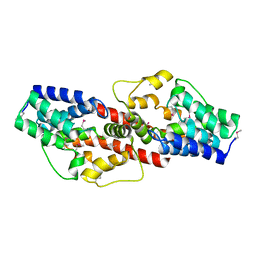 | | Crystal structure of Q88U62_LACPL from Lactobacillus plantarum. Northeast Structural Genomics target LpR71 | | Descriptor: | Uncharacterized protein lp_2664 | | Authors: | Benach, J, Su, M, Seetharaman, J, Forouhar, F, Chen, C.X, Cunningham, K, Ma, L.-C, Owens, L, Baran, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-16 | | Release date: | 2007-05-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Q88U62_LACPL from Lactobacillus plantarum.
To be Published
|
|
2Q88
 
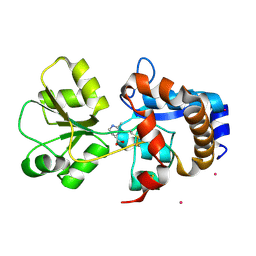 | | Crystal structure of EhuB in complex with ectoine | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
3DJU
 
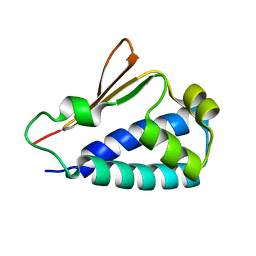 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
1G7D
 
 | | NMR STRUCTURE OF ERP29 C-DOMAIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, A, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
2O0M
 
 | | The crystal structure of the putative SorC family transcriptional regulator from Enterococcus faecalis | | Descriptor: | PHOSPHATE ION, Transcriptional regulator, SorC family | | Authors: | Zhang, R, Zhou, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-27 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the putative SorC family transcriptional regulator from Enterococcus faecalis
To be Published
|
|
2C5S
 
 | | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, PROBABLE THIAMINE BIOSYNTHESIS PROTEIN THII | | Authors: | Waterman, D.G, Ortiz-Lombardia, M, Fogg, M.J, Koonin, E.V, Antson, A.A. | | Deposit date: | 2005-11-01 | | Release date: | 2005-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain.
J.Mol.Biol., 356, 2006
|
|
2O5H
 
 | | Uncharacterized Protein Conserved in Bacteria, COG3792 from Neisseria meningitidis | | Descriptor: | Hypothetical protein | | Authors: | Kim, Y, Li, H, Gu, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-06 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Uncharacterized Protein Conserved in Bacteria, COG3792 from Neisseria meningitidis
To be Published
|
|
3DDV
 
 | | The crystal structure of the transcriptional regulator (GntR family) from Enterococcus faecalis V583 | | Descriptor: | MAGNESIUM ION, Transcriptional regulator (GntR family) | | Authors: | Zhang, R, Zhou, M, Bargassa, M, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-06 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of the transcriptional regulator (GntR family) from Enterococcus faecalis V583
To be Published
|
|
3DJN
 
 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
2VGO
 
 | | Crystal structure of Aurora B kinase in complex with Reversine inhibitor | | Descriptor: | INNER CENTROMERE PROTEIN A, N~6~-cyclohexyl-N~2~-(4-morpholin-4-ylphenyl)-9H-purine-2,6-diamine, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | D'Alise, A.M, Amabile, G, Iovino, M, Di Giorgio, F.P, Bartiromo, M, Sessa, F, Villa, F, Musacchio, A, Cortese, R. | | Deposit date: | 2007-11-15 | | Release date: | 2008-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reversine, a Novel Aurora Kinases Inhibitor, Inhibits Colony Formation of Human Acute Myeloid Leukemia Cells.
Mol.Cancer Ther., 7, 2008
|
|
3OQN
 
 | | Structure of ccpa-hpr-ser46-p-gntr-down cre | | Descriptor: | 5'-D(*AP*TP*GP*GP*TP*AP*CP*CP*GP*CP*TP*TP*TP*CP*AP*A)-3', 5'-D(*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*TP*AP*CP*CP*AP*T)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
4E1C
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mg++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
4DTD
 
 | | Structure and functional characterization of a Vibrio cholerae toxin from the MARTX/VgrG family. | | Descriptor: | GLYCEROL, VgrG protein | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and functional characterization of a Vibrio cholerae toxin
from the MARTX/VgrG family.
J.Biol.Chem., 2012
|
|
4DTL
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-02-21 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ATP and Mn++
J.Biol.Chem., 2012
|
|
4E1D
 
 | | Structure of a VgrG Vibrio cholerae toxin ACD domain in complex with ADP and Mn++ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Durand, E, Audoly, G, Derrez, E, Spinelli, S, Ortiz-Lombardia, M, Cascales, E, Raoult, D, Cambillau, C. | | Deposit date: | 2012-03-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and functional characterization of the Vibrio cholerae toxin
from the VgrG/MARTX family.
J.Biol.Chem., 2012
|
|
