6J4J
 
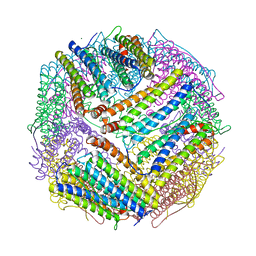 | | soybean seed H-2 ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Zhang, X, Zang, J, Chen, H, Zhao, G. | | Deposit date: | 2019-01-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Thermostability of protein nanocages: the effect of natural extra peptide on the exterior surface.
Rsc Adv, 9, 2019
|
|
6K98
 
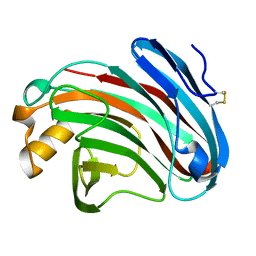 | | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family | | Descriptor: | GH12 beta-1, 4-endoglucanase | | Authors: | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, yingguo, B, Bin, Y. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
6K9D
 
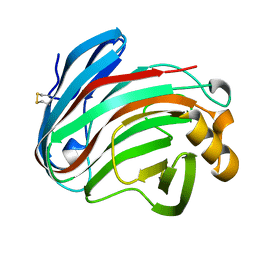 | | glycoside hydrolase family 12 (GH12) englucanase | | Descriptor: | GH12 beta-1, 4-endoglucanase | | Authors: | Hong, Y, Tao, T, Pengjun, S, Jiaming, C, Xiaoyu, W, Chen, H, Yingguo, B, Bin, Y. | | Deposit date: | 2019-06-14 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.505 Å) | | Cite: | Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
To Be Published
|
|
6JYW
 
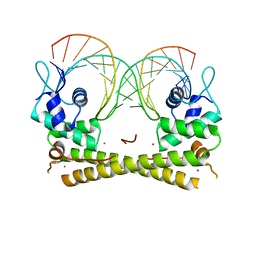 | |
7WPY
 
 | | AndA_M119A_N121V variant | | Descriptor: | Dioxygenase andA, FE (III) ION | | Authors: | Mori, T, Chen, H, Abe, I. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AndA_M119A_N121V variant
To Be Published
|
|
1PQN
 
 | | dominant negative human hDim1 (hDim1 1-128) | | Descriptor: | Spliceosomal U5 snRNP-specific 15 kDa protein | | Authors: | Zhang, Y.Z, Cheng, H, Gould, K.L, Golemis, E.A, Roder, H. | | Deposit date: | 2003-06-18 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure, stability and function of hDim1 investigated by NMR, circular
dichroism and mutational analysis
Biochemistry, 42, 2003
|
|
2A3D
 
 | | SOLUTION STRUCTURE OF A DE NOVO DESIGNED SINGLE CHAIN THREE-HELIX BUNDLE (A3D) | | Descriptor: | PROTEIN (DE NOVO THREE-HELIX BUNDLE) | | Authors: | Walsh, S.T.R, Cheng, H, Bryson, J.W, Roder, H, Degrado, W.F. | | Deposit date: | 1999-04-01 | | Release date: | 1999-05-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a de novo designed three-helix bundle protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2KZ2
 
 | | Calmodulin, C-terminal domain, F92E mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2J8J
 
 | | Solution Structure of the A4 Domain of Blood Coagulation Factor XI | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2J8L
 
 | | FXI Apple 4 domain loop-out conformation | | Descriptor: | COAGULATION FACTOR XI | | Authors: | Samuel, D, Cheng, H, Riley, P.W, Canutescu, A.A, Bu, Z, Walsh, P.N, Roder, H. | | Deposit date: | 2006-10-25 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the A4 Domain of Factor Xi Sheds Light on the Mechanism of Zymogen Activation.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2M3S
 
 | | Calmodulin, i85l, f92e, h107i, l112r, a128t, m144r mutant | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Moroz, Y.S, Wu, Y, Cheng, H, Roder, H, Korendovych, I.V. | | Deposit date: | 2013-01-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A single mutation in a regulatory protein produces evolvable allosterically regulated catalyst of nonnatural reaction.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1J7A
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN D68K MUTANT | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, VanHooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollen, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1J7B
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E94K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1J7C
 
 | | STRUCTURE OF THE ANABAENA FERREDOXIN MUTANT E95K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN I | | Authors: | Hurley, J.K, Weber-Main, A.M, Stankovich, M.T, Benning, M.M, Thoden, J.B, Vanhooke, J.L, Holden, H.M, Chae, Y.K, Xia, B, Cheng, H, Markley, J.L, Martinez-Julvez, M, Gomez-Moreno, C, Schmeits, J.L, Tollin, G. | | Deposit date: | 2001-05-16 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
4HX1
 
 | | Structure of HLA-A68 complexed with a tumor antigen derived peptide | | Descriptor: | 9-mer peptide from Tyrosinase-related protein-2, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4I48
 
 | | Structure of HLA-A68 complexed with an HIV Env derived peptide | | Descriptor: | 9-mer peptide from Envelope glycoprotein gp160, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
4HWZ
 
 | | Structure of HLA-A68 complexed with an HIV derived peptide | | Descriptor: | 9-mer peptide from Pol protein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Niu, L, Cheng, H, Zhang, S, Tan, S, Zhang, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2012-11-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural basis for the differential classification of HLA-A*6802 and HLA-A*6801 into the A2 and A3 supertypes
Mol.Immunol., 55, 2013
|
|
8GTN
 
 | |
2J8P
 
 | | NMR structure of C-terminal domain of human CstF-64 | | Descriptor: | CLEAVAGE STIMULATION FACTOR 64 KDA SUBUNIT | | Authors: | Qu, X, Perez-Canadillas, J.M, Agrawal, S, De Baecke, J, Cheng, H, Varani, G, Moore, C. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domains of Vertebrate Cstf-64 and its Yeast Orthologue RNA15 Form a New Structure Critical for Mrna 3'-End Processing.
J.Biol.Chem., 282, 2007
|
|
6J2E
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP1 | | Descriptor: | Beta-2-microglobulin, EBOV-NP1, MHC class I antigen | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2G
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Ebola virus-derived peptide EBOV-NP2 | | Descriptor: | Beta-2-microglobulin, EBOV-NP2, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2H
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 mutant (Met52 Asp53 Leu54 deleted) in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | Beta-2-microglobulin, HeV1, Ptal-N*01:01 (Met52 Asp53 Leu54 deleted) | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2D
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV1 | | Descriptor: | HeV1, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2F
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with Hendra virus-derived peptide HeV2 | | Descriptor: | HeV2, Ptal-N*01:01, beta-2 microglobulin | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
6J2I
 
 | | Crystal structure of bat (Pteropus Alecto) MHC class I Ptal-N*01:01 in complex with H17N10 influenza-like virus-derivrd peptide H17N10-NP | | Descriptor: | Beta-2-microglobulin, H17N10-NP, Ptal-N*01:01 | | Authors: | Lu, D, Liu, K.F, Yue, C, Lu, Q, Cheng, H, Chai, Y, Qi, J.X, Gao, G.F, Liu, W.J. | | Deposit date: | 2019-01-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Peptide presentation by bat MHC class I provides new insight into the antiviral immunity of bats.
Plos Biol., 17, 2019
|
|
