8HHT
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Hit-1 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]benzamide | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
8HHU
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with SY110 | | Descriptor: | (1~{R})-3,3-bis(fluoranyl)-~{N}-[(2~{R})-3-methoxy-1-oxidanylidene-1-[[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(1,3-thiazol-2-ylmethylamino)butan-2-yl]amino]propan-2-yl]cyclohexane-1-carboxamide, 3C-like proteinase nsp5 | | Authors: | Zeng, R, Xie, L.W, Huang, C, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | Deposit date: | 2022-11-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.258 Å) | | Cite: | A new generation M pro inhibitor with potent activity against SARS-CoV-2 Omicron variants.
Signal Transduct Target Ther, 8, 2023
|
|
5N0L
 
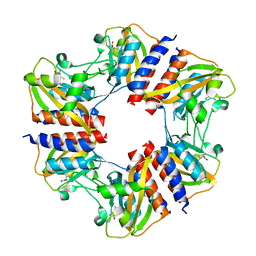 | | The structure of the cofactor binding GAF domain of the nutrient sensor CodY from Clostridium difficile | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, ISOLEUCINE | | Authors: | Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Sonenshein, A.L. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Impact of CodY protein on metabolism, sporulation and virulence in Clostridioides difficile ribotype 027.
Plos One, 14, 2019
|
|
7CHE
 
 | |
8HE4
 
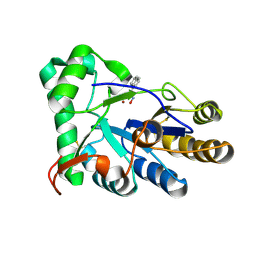 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
7CH4
 
 | |
5QCM
 
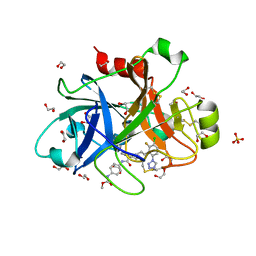 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl ~{N}-[4-[[(1~{S})-2-[(~{E})-3-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]phenyl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
6LK1
 
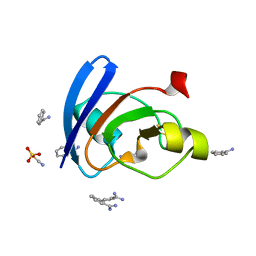 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | Descriptor: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
8HE2
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
 | |
6KV0
 
 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | Descriptor: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Onishi, Y, Kurisu, G, Tanaka, H. | | Deposit date: | 2019-09-02 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
5QCL
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
7CHB
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with BD-236 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BD-236 Fab heavy chain, BD-236 Fab light chain, ... | | Authors: | Xiao, J, Zhu, Q. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structurally Resolved SARS-CoV-2 Antibody Shows High Efficacy in Severely Infected Hamsters and Provides a Potent Cocktail Pairing Strategy.
Cell, 183, 2020
|
|
6ITX
 
 | |
7CHC
 
 | |
7CH5
 
 | |
5QCN
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-[[(1~{S})-2-[(~{E})-3-[5-chloranyl-2-(1,2,3,4-tetrazol-1-yl)phenyl]prop-2-enoyl]-5-[(3~{S})-3-ethoxycarbonylpiperidin-1-yl]carbonyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]carbonylamino]benzoic acid, Coagulation factor XI, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-08 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212).
J. Med. Chem., 60, 2017
|
|
4IN0
 
 | |
5KKF
 
 | |
4GHU
 
 | | Crystal structure of TRAF3/Cardif | | Descriptor: | Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 3 | | Authors: | Zhang, P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
5HN7
 
 | |
