5VSQ
 
 | | ABA-mimicking ligand AMF2beta in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(3,5-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.618 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
6DMK
 
 | | A multiconformer ligand model of an isoxazolyl-benzimidazole ligand bound to the bromodomain of human CREBBP | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-(2-phenylethyl)-1H-benzimidazole, CREB-binding protein, ... | | Authors: | Hudson, B.M, van Zundert, G, Keedy, D.A, Fonseca, R, Heliou, A, Suresh, P, Borrelli, K, Day, T, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | qFit-ligand Reveals Widespread Conformational Heterogeneity of Drug-Like Molecules in X-Ray Electron Density Maps.
J. Med. Chem., 61, 2018
|
|
2OPM
 
 | | Human Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 | | Descriptor: | 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, Farnesyl pyrophosphate synthetase (FPP synthetase) (FPS) (Farnesyl diphosphate synthetase) [Includes: Dimethylallyltranstransferase (EC 2.5.1.1); Geranyltranstransferase (EC 2.5.1.10)], MAGNESIUM ION, ... | | Authors: | Cao, R, Gao, Y.G, Robinson, H, Goddard, A. | | Deposit date: | 2007-01-29 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
2ZML
 
 | | Crystal structure of basic winged bean lectin in complex with Gal-ALPHA 1,4 Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7CIM
 
 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | Descriptor: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CL5
 
 | | The crystal structure of KanJ in complex with kanamycin B and N-oxalylglycine | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, Kanamycin B dioxygenase, N-OXALYLGLYCINE, ... | | Authors: | Kitayama, Y, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2020-07-20 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stepwise Post-glycosylation Modification of Sugar Moieties in Kanamycin Biosynthesis.
Chembiochem, 22, 2021
|
|
6HUP
 
 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with diazepam (Valium), GABA and megabody Mb38. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-CHLORO-1-METHYL-5-PHENYL-1,3-DIHYDRO-2H-1,4-BENZODIAZEPIN-2-ONE, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
1A2C
 
 | | Structure of thrombin inhibited by AERUGINOSIN298-A from a BLUE-GREEN ALGA | | Descriptor: | Aeruginosin 298-A, Hirudin variant-2, SODIUM ION, ... | | Authors: | Rios-Steiner, J.L, Murakami, M, Tulinsky, A. | | Deposit date: | 1997-12-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Thrombin Inhibited by Aeruginosin 298-A from a Blue-Green Alga
J.Am.Chem.Soc., 120, 1998
|
|
5VVI
 
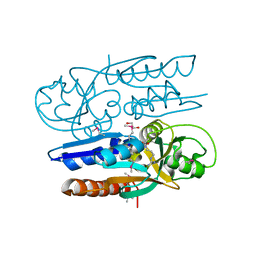 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
6HZ9
 
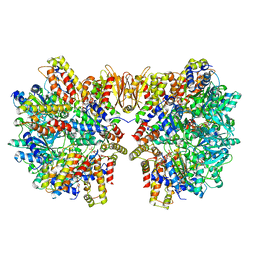 | | Structure of McrBC without DNA binding domains (Class 5) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Nirwan, N, Saikrishnan, K, Amunts, A. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC.
Nat Commun, 10, 2019
|
|
5VQD
 
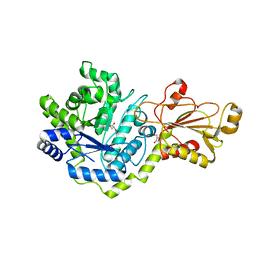 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
1ABI
 
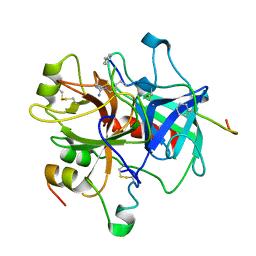 | |
6I0P
 
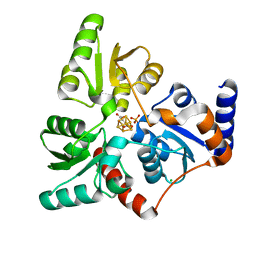 | | Structure of quinolinate synthase in complex with 6-mercaptopyridine-2,3-dicarboxylic acid | | Descriptor: | 6-mercaptopyridine-2,3-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
3GK7
 
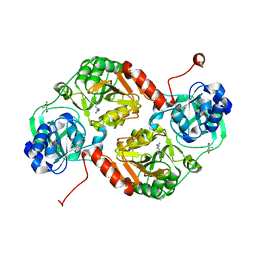 | | Crystal structure of 4-hydroxybutyrate CoA-Transferase from Clostridium aminobutyricum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-Hydroxybutyrate CoA-transferase, SPERMIDINE | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M. | | Deposit date: | 2009-03-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum
Biol.Chem., 390, 2009
|
|
5VS5
 
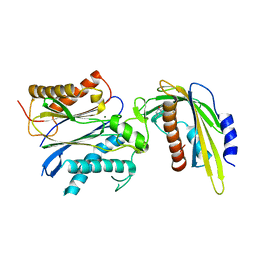 | | ABA-mimicking ligand AMF2alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(2,3-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-W, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
7CTR
 
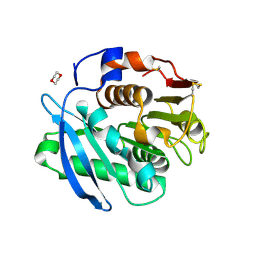 | | Closed form of PET-degrading cutinase Cut190 with thermostability-improving mutations of S226P/R228S/Q138A/D250C-E296C/Q123H/N202H | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Alpha/beta hydrolase family protein | | Authors: | Emori, M, Numoto, N, Senga, A, Bekker, G.J, Kamiya, N, Ito, N, Kawai, F, Oda, M. | | Deposit date: | 2020-08-20 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.
Proteins, 89, 2021
|
|
6HY3
 
 | | Three-dimensional structure of AgaC from Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, Beta-agarase C, GLYCEROL, ... | | Authors: | Naretto, A, Fanuel, M, Ropartz, D, Rogniaux, H, Larocque, R, Czjzek, M, Tellier, C, Michel, G. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The agar-specific hydrolaseZgAgaC from the marine bacteriumZobellia galactanivoransdefines a new GH16 protein subfamily.
J.Biol.Chem., 294, 2019
|
|
7C2P
 
 | | Structure of Egk Peptide | | Descriptor: | Plant defensing Egk | | Authors: | El Sahili, A. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Acs Pharmacol Transl Sci, 3, 2020
|
|
1AMY
 
 | |
2OW0
 
 | | MMP-9 active site mutant with iodine-labeled carboxylate inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix metalloproteinase-9 (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB), ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
6HWI
 
 | | Immature M-PMV capsid hexamer structure in intact virus particles | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Qu, K, Glass, B, Dolezal, M, Schur, F.K.M, Rein, A, Rumlova, M, Ruml, T, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HZ7
 
 | | Structure of McrBC without DNA binding domains (Class 3) | | Descriptor: | 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Nirwan, N, Saikrishnan, K, Amunts, A. | | Deposit date: | 2018-10-22 | | Release date: | 2019-07-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC.
Nat Commun, 10, 2019
|
|
4E8O
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ih from Acinetobacter baumannii | | Descriptor: | Aac(6')-Ih protein, CHLORIDE ION | | Authors: | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
6HWW
 
 | | Immature MLV capsid hexamer structure in intact virus particles | | Descriptor: | Putative gag polyprotein | | Authors: | Qu, K, Glass, B, Dolezal, M, Schur, F.K.M, Rein, A, Rumlova, M, Ruml, T, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2018-10-15 | | Release date: | 2018-12-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and architecture of immature and mature murine leukemia virus capsids.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W27
 
 | | Crystal structure of TnmS3 in complex with tiancimycin (TNM B) | | Descriptor: | Glyoxalase/bleomycin resisance protein/dioxygenase, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Chang, C.Y, Chang, C, Nocek, B, Rudolf, J.D, Joachimiak, A, Phillips Jr, G.N, SHen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-06-05 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
To Be Published
|
|
